3KSD
 
 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.2 angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-22 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
5HSH
 
 | |
6ALZ
 
 | | Crystal structure of Protein Phosphatase 1 bound to the natural inhibitor Tautomycetin | | 分子名称: | (2Z)-2-[(1R)-3-{[(2R,3S,4R,7S,8S,11S,13R,16E)-17-ethyl-4,8-dihydroxy-3,7,11,13-tetramethyl-6,15-dioxononadeca-16,18-dien-2-yl]oxy}-1-hydroxy-3-oxopropyl]-3-methylbut-2-enedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2017-08-08 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.208 Å) | | 主引用文献 | PP1:Tautomycetin Complex Reveals a Path toward the Development of PP1-Specific Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
6H13
 
 | | Crystal structure of TcACHE complexed to1-(4-((Methyl((1-(2-((1,2,3,4-tetrahydroacridin-9-yl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)pyridin-2-yl)-3-(6-oxo-1,2,3,4,6,10b-hexahydropyrido[2,1-a]isoindol-10-yl)urea | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Coquelle, N, Colletier, J.P. | | 登録日 | 2018-07-10 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Design, biological evaluation and X-ray crystallography of nanomolar multifunctional ligands targeting simultaneously acetylcholinesterase and glycogen synthase kinase-3.
Eur.J.Med.Chem., 168, 2019
|
|
4P2M
 
 | | Swapped Dimer of Mycobacterial Adenylyl cyclase Rv1625c: Form 1 | | 分子名称: | Adenylate cyclase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | 著者 | Barathy, D.V, Mattoo, R, Visweswariah, S.S, Suguna, K. | | 登録日 | 2014-03-04 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | New structural forms of a mycobacterial adenylyl cyclase Rv1625c.
Iucrj, 1, 2014
|
|
6AT8
 
 | |
6F35
 
 | | Crystal structure of the aspartate aminotranferase from Rhizobium meliloti | | 分子名称: | ACETATE ION, Aspartate aminotransferase B, GLYCEROL, ... | | 著者 | Cobessi, D, Graindorge, M, Giustini, C, Matringe, M. | | 登録日 | 2017-11-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
5NGL
 
 | | The endo-beta1,6-glucanase BT3312 | | 分子名称: | Glucosylceramidase, SODIUM ION, beta-D-glucopyranose-(1-6)-1-DEOXYNOJIRIMYCIN | | 著者 | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | 登録日 | 2017-03-17 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
5NMB
 
 | | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain | | 分子名称: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-1-[(2~{S},5~{R})-5-(2-oxidanylpropan-2-yl)oxolan-2-yl]ethyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Belorusova, A.Y. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain.
Eur J Med Chem, 134, 2017
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | 著者 | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-09-27 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
6H70
 
 | |
3L6O
 
 | | Crystal Structure of Phosphate bound apo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.2 Angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, PHOSPHATE ION | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-12-23 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
5XDQ
 
 | | Bovine heart cytochrome c oxidase in the fully oxidized state with pH 7.3 at 1.77 angstrom resolution | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | 登録日 | 2017-03-29 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structure of bovine cytochrome c oxidase crystallized at a neutral pH using a fluorinated detergent.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6FFM
 
 | |
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | 分子名称: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | 著者 | Robin, A.Y, Girard, E, Madern, D. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
5NMA
 
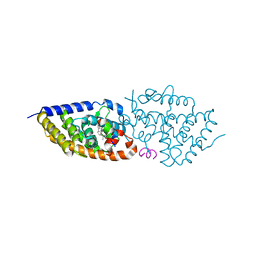 | | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain | | 分子名称: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-1-[(2~{S},5~{S})-5-(2-oxidanylpropan-2-yl)oxolan-2-yl]ethyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Belorusova, A.Y. | | 登録日 | 2017-04-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-activity relationship study of vitamin D analogs with oxolane group in their side chain.
Eur J Med Chem, 134, 2017
|
|
3ENK
 
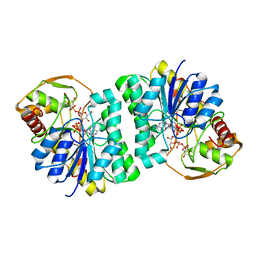 | |
3LVF
 
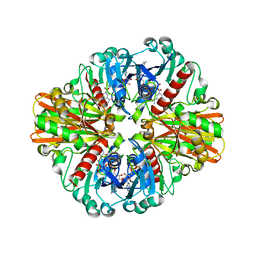 | |
7A1Q
 
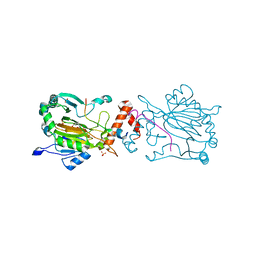 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | 分子名称: | 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Brewitz, L, Schofield, C.J. | | 登録日 | 2020-08-13 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7Y3F
 
 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | 登録日 | 2022-06-10 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
5JCB
 
 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | 分子名称: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Guan, Z, Zhao, W, Yin, P. | | 登録日 | 2016-04-14 | | 公開日 | 2017-09-27 | | 最終更新日 | 2025-09-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | 分子名称: | Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5ZLZ
 
 | | Structure of tPA and PAI-1 | | 分子名称: | GLYCEROL, Plasminogen activator inhibitor 1, Tissue-type plasminogen activator | | 著者 | Min, L, Huang, M. | | 登録日 | 2018-03-31 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.581 Å) | | 主引用文献 | Development of a PAI-1 trapping agent (PAItrap2) based on inactivated tPA-SPD and the crystal structure of PAItrap2 in complex with PAI-1
To Be Published
|
|
5EPS
 
 | |
7QSL
 
 | | Bovine complex I in lipid nanodisc, Active-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
