2VW5
 
 | | Structure Of The Hsp90 Inhibitor 7-O-carbamoylpremacbecin Bound To The N- Terminus Of Yeast Hsp90 | | 分子名称: | (4E,8S,9R,10E,12S,13R,14S,16R)-13,20-dihydroxy-14-methoxy-4,8,10,12,16-pentamethyl-3-oxo-2-azabicyclo[16.3.1]docosa-1(22),4,10,18,20-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, SULFATE ION | | 著者 | Roe, S.M, Prodromou, C, Pearl, L.H. | | 登録日 | 2008-06-16 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Optimizing Natural Products by Biosynthetic Engineering: Discovery of Nonquinone Hsp90 Inhibitors.
J.Med.Chem., 51, 2008
|
|
3UB8
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with formamide bound | | 分子名称: | FORMAMIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | 登録日 | 2011-10-23 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3TIQ
 
 | | Crystal structure of Staphylococcus aureus SasG G51-E-G52 module | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Surface protein G | | 著者 | Gruszka, D.T, Wojdyla, J.A, Turkenburg, J.P, Potts, J.R. | | 登録日 | 2011-08-21 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8739 Å) | | 主引用文献 | Staphylococcal biofilm-forming protein has a contiguous rod-like structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UB9
 
 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with hydroxyurea bound | | 分子名称: | GLYCEROL, N-HYDROXYUREA, SULFATE ION, ... | | 著者 | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | 登録日 | 2011-10-23 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | 分子名称: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | 著者 | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | 登録日 | 2011-07-20 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
2LDI
 
 | |
5V1L
 
 | | Structure of S-GNA dodecamer | | 分子名称: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*(ZTH)P*AP*GP*CP*G)-3'), SPERMINE, STRONTIUM ION | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2017-03-02 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
5V1K
 
 | | Structure of R-GNA dodecamer | | 分子名称: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*(5BU)P*(8RJ)P*AP*GP*CP*G)-3') | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2017-03-02 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 2017
|
|
3V1K
 
 | |
3V3D
 
 | |
3V1N
 
 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | 分子名称: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | 著者 | Ghosh, S, Bolin, J.T. | | 登録日 | 2011-12-09 | | 公開日 | 2012-03-21 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3V1L
 
 | |
5VR4
 
 | | RNA octamer containing 2'-F-4'-OMe U. | | 分子名称: | COBALT TETRAAMMINE ION, RNA (5'-R(*CP*GP*AP*AP*(UMO)P*UP*CP*G)-3') | | 著者 | Harp, J.M, Egli, M. | | 登録日 | 2017-05-10 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 4'-C-Methoxy-2'-deoxy-2'-fluoro Modified Ribonucleotides Improve Metabolic Stability and Elicit Efficient RNAi-Mediated Gene Silencing.
J. Am. Chem. Soc., 139, 2017
|
|
6ZP5
 
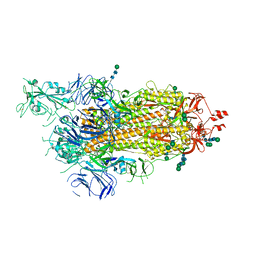 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Martinez, M, Marabini, R, Carazo, J.M. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
2MH8
 
 | | GA-79-MBP cs-rosetta structures | | 分子名称: | GA-79-MBP, maltose binding protein | | 著者 | He, Y, Chen, Y, Porter, L, Bryan, P, Orban, J. | | 登録日 | 2013-11-19 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Subdomain interactions foster the design of two protein pairs with 80% sequence identity but different folds.
Biophys.J., 108, 2015
|
|
5V2H
 
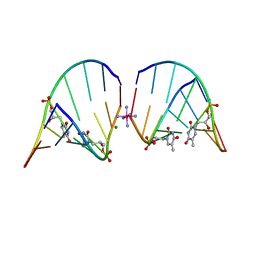 | | RNA octamer containing glycol nucleic acid, SgnT | | 分子名称: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*(CBV)P*GP*AP*AP*(ZTH)P*UP*CP*G)-3') | | 著者 | Harp, J.M, Egli, M. | | 登録日 | 2017-03-04 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.080013 Å) | | 主引用文献 | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
3V1M
 
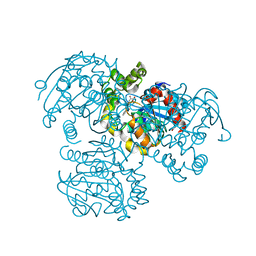 | | Crystal Structure of the S112A/H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | 分子名称: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION | | 著者 | Ghosh, S, Bolin, J.T. | | 登録日 | 2011-12-09 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
6ZP7
 
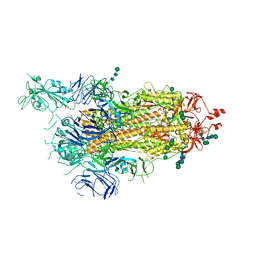 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Martinez, M, Marabini, R, Carazo, J.M. | | 登録日 | 2020-07-08 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZOW
 
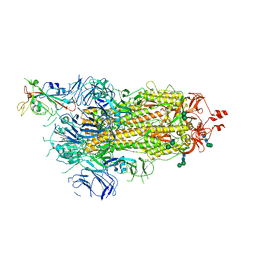 | | SARS-CoV-2 spike in prefusion state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Martinez, M, Marabini, R, Carazo, J.M. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
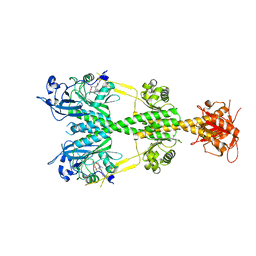
3TCT
 
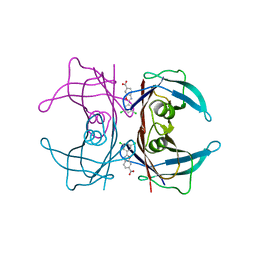 | | Structure of wild-type TTR in complex with tafamidis | | 分子名称: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | 著者 | Connelly, S, Kelly, J.W, Wilson, I.A. | | 登録日 | 2011-08-09 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5UYR
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant D199A from Xanthomonas campestris | | 分子名称: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | 著者 | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | 登録日 | 2017-02-24 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
2MKX
 
 | |
3UB6
 
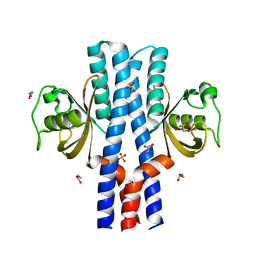 | | Periplasmic portion of the Helicobacter pylori chemoreceptor TlpB with urea bound | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | 著者 | Henderson, J.N, Sweeney, E.G, Goers, J, Wreden, C, Hicks, K.G, Parthasarathy, R, Guillemin, K.J, Remington, S.J. | | 登録日 | 2011-10-23 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB.
Structure, 20, 2012
|
|
4V1F
 
 | |
4V1G
 
 | |
