7OSB
 
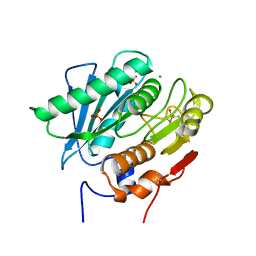 | | Crystal Structure of a Double Mutant PETase (S238F/W159H) from Ideonella sakaiensis | | 分子名称: | CHLORIDE ION, GLYCEROL, Poly(ethylene terephthalate) hydrolase, ... | | 著者 | Shakespeare, T.J, Zahn, M, Allen, M.D, McGeehan, J.E. | | 登録日 | 2021-06-08 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Comparative Performance of PETase as a Function of Reaction Conditions, Substrate Properties, and Product Accumulation.
ChemSusChem, 15, 2022
|
|
4U5L
 
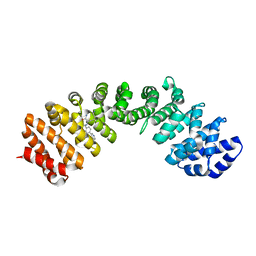 | |
4U5V
 
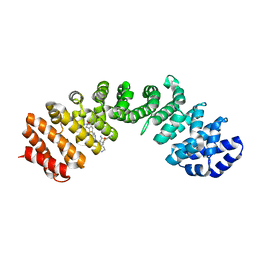 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | 分子名称: | Importin subunit alpha-1, N~2~-{[5-(pyridin-3-yl)thiophen-2-yl]methyl}-L-lysinamide | | 著者 | Stewart, M, Valkov, E, Holvey, R.S. | | 登録日 | 2014-07-25 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.968 Å) | | 主引用文献 | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U9C
 
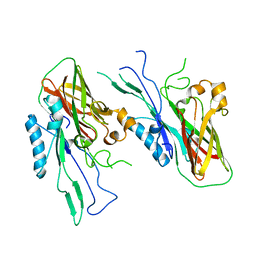 | |
4WUZ
 
 | |
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | 分子名称: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.593 Å) | | 主引用文献 | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAD
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | 分子名称: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | 著者 | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-07-02 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
6OAA
 
 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | 著者 | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
4U1Z
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form D | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9401 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U22
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form D | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4409 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U23
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form F | | 分子名称: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6734 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U5N
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | 分子名称: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysyl-N-[(1R,2S,3R)-1-{[(2R)-1-amino-1-oxo-3-phenylpropan-2-yl]amino}-1,3-dihydroxybutan-2-yl]glycinamide | | 著者 | Stewart, M, Valkov, E, Holvey, R.S. | | 登録日 | 2014-07-25 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5S
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | 分子名称: | Importin subunit alpha-1, N-[(2S)-2-[(N~2~-acetyl-D-lysyl)amino]-3-(pyridin-3-ylmethoxy)propyl]-L-allothreonyl-D-phenylalaninamide | | 著者 | Stewart, M, Valkov, E, Holvey, R.S. | | 登録日 | 2014-07-25 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
7MMN
 
 | |
4UCU
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | 著者 | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | 登録日 | 2014-12-04 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4WG4
 
 | |
4TYI
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 | | 分子名称: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, Fibroblast growth factor receptor 4 | | 著者 | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | 登録日 | 2014-07-08 | | 公開日 | 2014-09-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | 分子名称: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | 著者 | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
8ERM
 
 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | 分子名称: | B-type flagellin, GLYCEROL, SULFATE ION | | 著者 | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | 登録日 | 2022-10-12 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.475 Å) | | 主引用文献 | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
8F0G
 
 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | 著者 | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | 登録日 | 2022-11-02 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | 著者 | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | 登録日 | 2022-11-02 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8EMM
 
 | |
4U1O
 
 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | 著者 | Chen, L, Gouaux, E. | | 登録日 | 2014-07-15 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8501 Å) | | 主引用文献 | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
5KSE
 
 | | Flap endonuclease 1 (FEN1) R100A with 5'-flap substrate DNA and Sm3+ | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | 著者 | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | 登録日 | 2016-07-08 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
