3R4A
 
 | |
2PVC
 
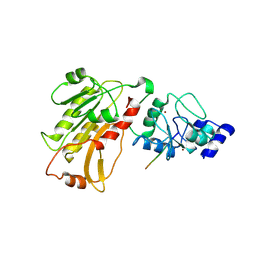 | | DNMT3L recognizes unmethylated histone H3 lysine 4 | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, Histone H3 peptide, ZINC ION | | 著者 | Cheng, X. | | 登録日 | 2007-05-09 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.69 Å) | | 主引用文献 | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
1AJK
 
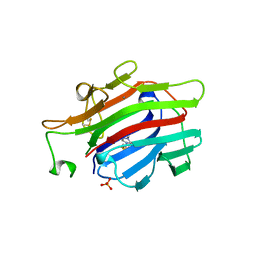 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | 著者 | Ay, J, Heinemann, U. | | 登録日 | 1997-05-06 | | 公開日 | 1998-05-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
3R48
 
 | | Crystal structure of a hetero-hexamer coiled coil | | 分子名称: | GLYCEROL, coiled coil helix W22-L24H, coiled coil helix Y15-L24D | | 著者 | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | 登録日 | 2011-03-17 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.0011 Å) | | 主引用文献 | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
1AJO
 
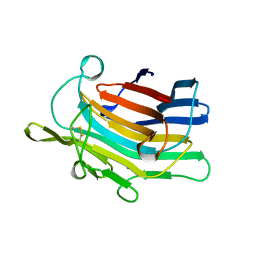 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | 分子名称: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | 著者 | Ay, J, Heinemann, U. | | 登録日 | 1997-05-07 | | 公開日 | 1998-05-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
2PV0
 
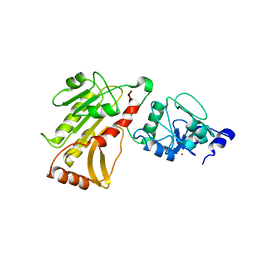 | | DNA methyltransferase 3 like protein (DNMT3L) | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, ZINC ION | | 著者 | Cheng, X. | | 登録日 | 2007-05-09 | | 公開日 | 2007-08-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA.
Nature, 448, 2007
|
|
3B3O
 
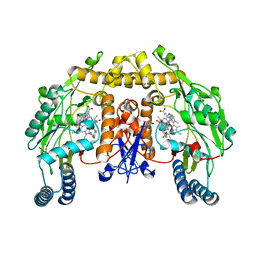 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2007-10-22 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3B3P
 
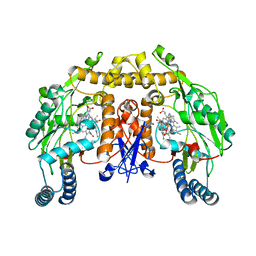 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2007-10-22 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
4E8U
 
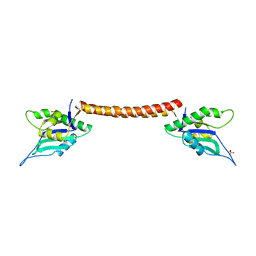 | |
2N2U
 
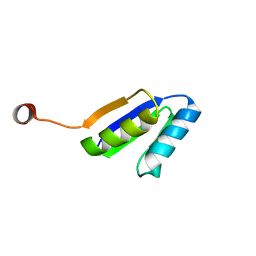 | | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358 | | 分子名称: | OR358 | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Hamilton, K, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-05-14 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of DE NOVO DESIGNED Ferredoxin Fold PROTEIN sfr3, Northeast Structural Genomics Consortium (NESG) Target OR358
To be Published
|
|
3S70
 
 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO solved by As-SAD | | 分子名称: | ACETATE ION, CACODYLATE ION, Caspase-6, ... | | 著者 | Su, X.-D, Liu, X, Wang, X.-J. | | 登録日 | 2011-05-26 | | 公開日 | 2012-04-11 | | 最終更新日 | 2012-12-12 | | 実験手法 | X-RAY DIFFRACTION (1.625 Å) | | 主引用文献 | Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer
Plos One, 6, 2011
|
|
3S2S
 
 | |
2QRV
 
 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Jia, D, Cheng, X. | | 登録日 | 2007-07-29 | | 公開日 | 2007-12-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
3U19
 
 | | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | 分子名称: | GLYCEROL, artificial protein OR51 | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-29 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE
ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
3DQS
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2008-07-09 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQT
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{trans-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2008-07-09 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQR
 
 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2008-07-09 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | 分子名称: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | 著者 | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | 登録日 | 2020-04-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7TEP
 
 | |
7SU2
 
 | |
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | 分子名称: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | 著者 | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | 登録日 | 2006-06-21 | | 公開日 | 2006-11-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
2NBS
 
 | |
4P6L
 
 | |
1EDQ
 
 | |
1EKY
 
 | |
