5LYU
 
 | | The native crystal structure of 7SK 5'-hairpin | | 分子名称: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | 登録日 | 2016-09-28 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
5BS7
 
 | | Structure of histone H3/H4 in complex with Spt2 | | 分子名称: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | 著者 | Chen, S, Patel, D.J. | | 登録日 | 2015-06-01 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
8QFC
 
 | | UFL1 E3 ligase bound 60S ribosome | | 分子名称: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | 著者 | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | 分子名称: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | 著者 | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | 登録日 | 2022-08-26 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.01 Å) | | 主引用文献 | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
7TYR
 
 | |
7NFC
 
 | | Cryo-EM structure of NHEJ super-complex (dimer) | | 分子名称: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-05 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
 | | Cryo-EM structure of NHEJ super-complex (monomer) | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2021-02-06 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
2JR0
 
 | | Solution structure of NusB from Aquifex Aeolicus | | 分子名称: | N utilization substance protein B homolog | | 著者 | Das, R, Loss, S, Li, J, Tarasov, S, Wingfield, P, Waugh, D.S, Byrd, R.A, Altieri, A.S. | | 登録日 | 2007-06-18 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural biophysics of the NusB:NusE antitermination complex.
J.Mol.Biol., 376, 2008
|
|
6PY8
 
 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | 分子名称: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | 著者 | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | 登録日 | 2019-07-29 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.75 Å) | | 主引用文献 | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
3FEY
 
 | |
3FEX
 
 | |
8BOT
 
 | |
8BFC
 
 | | Binary structure of 14-3-3s and RND3 phosphopeptide | | 分子名称: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Somsen, B.A, Ottmann, C. | | 登録日 | 2022-10-24 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Reversible Dual-Covalent Molecular Locking of the 14-3-3/ERR gamma Protein-Protein Interaction as a Molecular Glue Drug Discovery Approach.
J.Am.Chem.Soc., 145, 2023
|
|
7PPZ
 
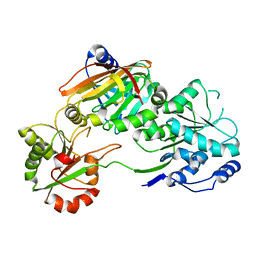 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form A | | 分子名称: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | 著者 | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | 登録日 | 2021-09-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7PQ0
 
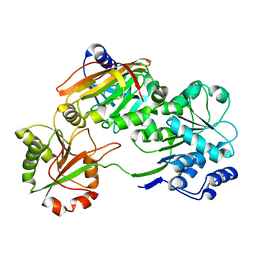 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) C94S inactive mutant in complex with human eIF4A - Crystal form B | | 分子名称: | Burkholderia Lethal Factor 1 (BLF1), Eukaryotic initiation factor 4A-I | | 著者 | Mobbs, G.W, Aziz, A.A, Dix, S.R, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | 登録日 | 2021-09-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
7XJG
 
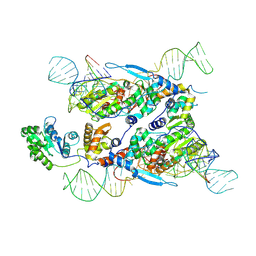 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | 分子名称: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | 著者 | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | 登録日 | 2022-04-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
4ID5
 
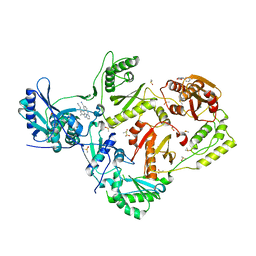 | | HIV-1 reverse transcriptase with bound fragment at the RNase H primer grip site | | 分子名称: | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | 著者 | Bauman, J.D, Patel, D, Arnold, E. | | 登録日 | 2012-12-11 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4GMJ
 
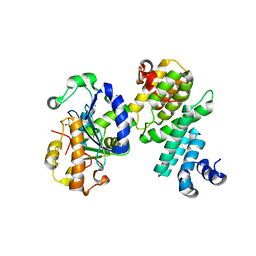 | | Structure of human NOT1 MIF4G domain co-crystallized with CAF1 | | 分子名称: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 7, CHLORIDE ION, ... | | 著者 | Petit, P, Weichenrieder, O, Wohlbold, L, Izaurralde, E. | | 登録日 | 2012-08-16 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex
Nucleic Acids Res., 40, 2012
|
|
1H2U
 
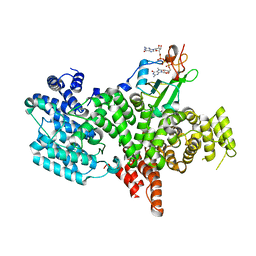 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | 分子名称: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | 著者 | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | 登録日 | 2002-08-16 | | 公開日 | 2002-10-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2T
 
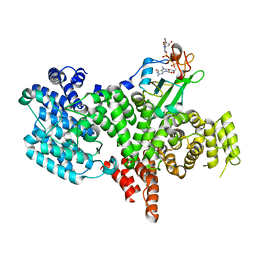 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | 分子名称: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | 著者 | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | 登録日 | 2002-08-16 | | 公開日 | 2002-10-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
1H2V
 
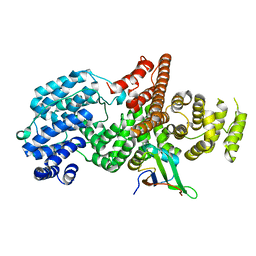 | | Structure of the human nuclear cap-binding-complex (CBC) | | 分子名称: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 80 KDA NUCLEAR CAP BINDING PROTEIN | | 著者 | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | 登録日 | 2002-08-16 | | 公開日 | 2002-10-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
5F5O
 
 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | 分子名称: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | 著者 | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | 登録日 | 2015-12-04 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
7S4E
 
 | |
7SN8
 
 | |
7F5S
 
 | | human delta-METTL18 60S ribosome | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Takahashi, M, Kashiwagi, K, Ito, T. | | 登録日 | 2021-06-22 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
