8PKA
 
 | |
8PK9
 
 | |
8JMN
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-21 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[[2-[(4-chlorophenyl)methoxy]phenyl]methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abe, K, Yokoshima, S, Yoshimori, A. | | 登録日 | 2023-06-05 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
6F57
 
 | |
8JSM
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | 分子名称: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | 著者 | Qi, P, Yi, S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
8JSL
 
 | | The structure of EBOV L-VP35-RNA complex | | 分子名称: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV, ... | | 著者 | Qi, P, Yi, S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
5NNL
 
 | |
8JSN
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 2) | | 分子名称: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome, ... | | 著者 | Qi, P, Yi, S. | | 登録日 | 2023-06-20 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | 著者 | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | 登録日 | 2015-08-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5D9D
 
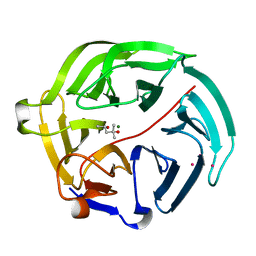 | | Luciferin-regenerating enzyme solved by SAD using synchrotron radiation at room temperature | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | 著者 | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | 登録日 | 2015-08-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5D9C
 
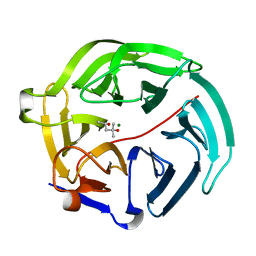 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against Hg derivative data) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | 著者 | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | 登録日 | 2015-08-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
6BRR
 
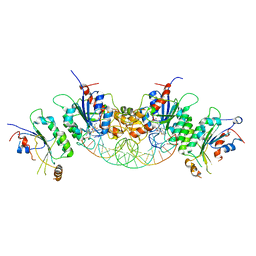 | |
4C11
 
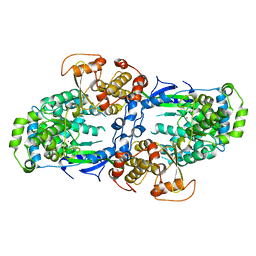 | |
1U0I
 
 | | IAAL-E3/K3 heterodimer | | 分子名称: | IAAL-E3, IAAL-K3 | | 著者 | Lindhout, D.A, Litowski, J.R, Mercier, P, Hodges, R.S, Sykes, B.D. | | 登録日 | 2004-07-13 | | 公開日 | 2004-10-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of a highly stable de novo heterodimeric coiled-coil
Biopolymers, 75, 2004
|
|
5K7F
 
 | | Crystal structure of apo AibR | | 分子名称: | ACETATE ION, Transcriptional regulator, TetR family | | 著者 | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | 登録日 | 2016-05-26 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
5CIY
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3'), Modification methylase HhaI, ... | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
5K7H
 
 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | 分子名称: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | 著者 | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | 登録日 | 2016-05-26 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
5CIU
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
5KEZ
 
 | |
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | 分子名称: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.95063877 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
5JD2
 
 | | SFX structure of corestreptavidin-selenobiotin complex | | 分子名称: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | 著者 | DeMirci, H, Hunter, M.S, Boutet, S. | | 登録日 | 2016-04-15 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
4ONJ
 
 | |
6U8P
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | 分子名称: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-05 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8V
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | 分子名称: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Zhang, Z.M, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
4ONQ
 
 | |
