1MMJ
 
 | | Porcine pancreatic elastase complexed with a potent peptidyl inhibitor, FR136706 | | 分子名称: | 2-[4-[[(S)-1-[[(S)-2-[[(RS)-3,3,3-TRIFLUORO-1-ISOPROPYL-2-OXOPROPYL]AMINOCARBONYL]PYRROLIDIN-1-YL-]CARBONYL]-2-METHYLPROPYL]AMINOCARBONYL]BENZOYLAMINO]ACETIC ACID, CALCIUM ION, SULFATE ION, ... | | 著者 | Kinoshita, T. | | 登録日 | 2002-09-04 | | 公開日 | 2002-12-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | True interaction mode of porcine pancreatic elastase with FR136706, a
potent peptidyl inhibitor
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1MMX
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-fucose | | 分子名称: | Aldose 1-epimerase, SODIUM ION, alpha-L-fucopyranose | | 著者 | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | 登録日 | 2002-09-04 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
3WB4
 
 | | Crystal Structure of beta secetase in complex with 2-amino-3,6-dimethyl-6-(2-phenylethyl)-3,4,5,6-tetrahydropyrimidin-4-one | | 分子名称: | (6R)-2-amino-3,6-dimethyl-6-(2-phenylethyl)-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | 著者 | Yonezawa, S, Fujiwara, K, Yamamoto, T, Hattori, K, Yamakawa, H, Muto, C, Hosono, M, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | 登録日 | 2013-05-13 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Conformational restriction approach to beta-secretase (BACE1) inhibitors III: Effective investigation of the binding mode by combinational use of X-ray analysis, isothermal titration calorimetry and theoretical calculations
Bioorg.Med.Chem., 21, 2013
|
|
5PAC
 
 | | human factor VIIa in complex with 5-hydroxy-N-(4-oxo-3H-quinazolin-6-yl)-1-[3-[(phenylcarbamoylamino)methyl]phenyl]pyrazole-4-carboxamide at 1.50A | | 分子名称: | 5-hydroxy-N-(4-oxo-3H-quinazolin-6-yl)-1-[3-[(phenylcarbamoylamino)methyl]phenyl]pyrazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | 登録日 | 2016-11-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of a Factor VIIa complex
To be published
|
|
3WCF
 
 | | The complex structure of HsSQS wtih ligand,BPH1218 | | 分子名称: | Squalene synthase, hydrogen [(1S)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | 著者 | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | 登録日 | 2013-05-27 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
5PAS
 
 | | Crystal Structure of Factor VIIa in complex with (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]-3-phenylpropanamide | | 分子名称: | (2S)-2-hydroxy-N-[[3-[5-hydroxy-4-(1H-pyrrolo[3,2-c]pyridin-2-yl)pyrazol-1-yl]phenyl]methyl]-3-phenylpropanamide, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | 登録日 | 2016-11-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal Structure of a Factor VIIa complex
To be published
|
|
5PBC
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B in complex with N09724a | | 分子名称: | 1,2-ETHANEDIOL, 4-bromo-1H-imidazole, Bromodomain adjacent to zinc finger domain protein 2B | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-03 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.771 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3HC6
 
 | | FXR with SRC1 and GSK088 | | 分子名称: | 3-[(5-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1H-indol-1-yl)methyl]benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | 著者 | Williams, S.P, Madauss, K.P. | | 登録日 | 2009-05-05 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2NN5
 
 | | Structure of Conserved Protein of Unknown Function EF2215 from Enterococcus faecalis | | 分子名称: | 1,2-ETHANEDIOL, Hypothetical protein EF_2215, MAGNESIUM ION | | 著者 | Osipiuk, J, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-10-23 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | X-ray crystal structure of conserved hypothetical protein EF_2215 from Enterococcus faecalis.
To be Published
|
|
7GUD
 
 | | Crystal Structure of B-cell lymphoma 6 protein BTB domain in complex with ligand 1 at 1.51 MGy X-ray dose. | | 分子名称: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | 著者 | Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | 登録日 | 2024-01-09 | | 公開日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
3WD3
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | 分子名称: | Chitinase B, GLYCEROL, SULFATE ION, ... | | 著者 | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | 登録日 | 2013-06-06 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5SPV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with REAL250003774401 | | 分子名称: | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WRL
 
 | |
253L
 
 | | LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Kuroki, R, Shoichet, B, Weaver, L.H, Matthews, B.W. | | 登録日 | 1997-11-10 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A relationship between protein stability and protein function.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2DEC
 
 | |
5SPI
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z4574659604 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{4-[(cyclopropanecarbonyl)amino]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1V66
 
 | | Solution structure of human p53 binding domain of PIAS-1 | | 分子名称: | Protein inhibitor of activated STAT protein 1 | | 著者 | Okubo, S, Hara, F, Tsuchida, Y, Shimotakahara, S, Suzuki, S, Hatanaka, H, Yokoyama, S, Tanaka, H, Yasuda, H, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-27 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers
J.Biol.Chem., 279, 2004
|
|
2JG4
 
 | | Substrate-free IDE structure in its closed conformation | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | 著者 | Malito, E, Tang, W.J. | | 登録日 | 2007-02-07 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
5SOI
 
 | |
7DDV
 
 | | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | 分子名称: | (1S)-1-(2-methyl-1,2,4-triazol-3-yl)ethanamine, CHLORIDE ION, GLYCEROL, ... | | 著者 | Tiwari, S, Pal, R.K, Biswal, B.K. | | 登録日 | 2020-10-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of M.tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor
To Be Published
|
|
1MD2
 
 | | CHOLERA TOXIN B-PENTAMER WITH DECAVALENT LIGAND BMSC-0013 | | 分子名称: | 3-ETHYLAMINO-4-METHYLAMINO-CYCLOBUTANE-1,2-DIONE, CHOLERA TOXIN B SUBUNIT, CYANIDE ION, ... | | 著者 | Zhang, Z, Merritt, E.A, Ahn, M, Roach, C, Hol, W.G.J, Fan, E. | | 登録日 | 2002-08-06 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin.
J.Am.Chem.Soc., 124, 2002
|
|
3WCJ
 
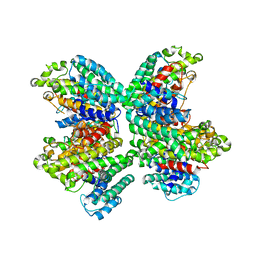 | | The complex structure of HsSQS wtih ligand,E5700 | | 分子名称: | (3R)-3-({2-benzyl-6-[(3R,4S)-3-hydroxy-4-methoxypyrrolidin-1-yl]pyridin-3-yl}ethynyl)-1-azabicyclo[2.2.2]octan-3-ol, Squalene synthase | | 著者 | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | 登録日 | 2013-05-27 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
4ESI
 
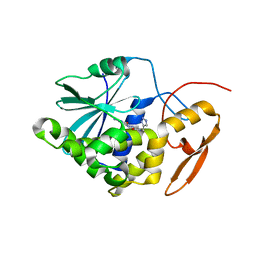 | | Structure of ricin A chain bound with N-((1H-1,2,3-triazol-4-yl)methyl-2-amino-4-oxo-3,4-dihydropteridine-7-carboxamide | | 分子名称: | 2-amino-4-oxo-N-(1H-1,2,3-triazol-5-ylmethyl)-1,4-dihydropteridine-7-carboxamide, Ricin | | 著者 | Jasheway, K.R, Pruet, J.M, Ryoto, S, Manzano, L.A, Wiget, P.A, Kamat, I, Anslyn, E.V, Monzingo, A.F, Robertus, J.D. | | 登録日 | 2012-04-23 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Optimized 5-membered heterocycle-linked pterins for the inhibition of Ricin Toxin A.
ACS Med Chem Lett, 3, 2012
|
|
4EPQ
 
 | |
5SE4
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cc(nc2nc(nn12)CCc3nc(cn3C)c4ccccc4)C5CC5)C, micromolar IC50=0.001357 | | 分子名称: | (8S)-5-cyclopropyl-7-methyl-2-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl][1,2,4]triazolo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | 著者 | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | 登録日 | 2022-01-21 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
