1UGU
 
 | | Crystal structure of PYP E46Q mutant | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Sugishima, M, Tanimoto, Y, Hamada, N, Tokunaga, F, Fukuyama, K. | | 登録日 | 2003-06-19 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6FCY
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ADP | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ATP phosphoribosyltransferase, ... | | 著者 | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | 登録日 | 2017-12-21 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
4RRV
 
 | |
6B90
 
 | | Multiconformer model of apo WT PTP1B with glycerol at 100 K (ALTERNATIVE REFINEMENT OF PDB 1SUG showing conformational heterogeneity) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | 登録日 | 2017-10-09 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1NE4
 
 | | Crystal Structure of Rp-cAMP Binding R1a Subunit of cAMP-dependent Protein Kinase | | 分子名称: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | 著者 | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | 登録日 | 2002-12-10 | | 公開日 | 2004-01-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
4RSS
 
 | | Crystal structure of tyrosine-protein kinase SYK with an inhibitor | | 分子名称: | 1-[(3-methyl-1-{2-[(1,2,3-trimethyl-1H-indol-5-yl)amino]pyrimidin-4-yl}-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | 著者 | Lee, B.I, Lee, S.J, Choi, J.-S. | | 登録日 | 2014-11-11 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Highly potent and selective pyrazolylpyrimidines as Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3RLL
 
 | | Crystal structure of the T877A androgen receptor ligand binding domain in complex with (S)-N-(4-Cyano-3-(trifluoromethyl)phenyl)-3-(4-cyanonaphthalen-1-yloxy)-2-hydroxy-2-methylpropanamide | | 分子名称: | (2S)-3-[(4-cyanonaphthalen-1-yl)oxy]-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydroxy-2-methylpropanamide, Androgen receptor | | 著者 | Bohl, C.E, Duke, C.B, Jones, A, Dalton, J.T, Miller, D.D. | | 登録日 | 2011-04-19 | | 公開日 | 2011-05-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Unexpected binding orientation of bulky-B-ring anti-androgens and implications for future drug targets.
J.Med.Chem., 54, 2011
|
|
7GPU
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with POB0122 | | 分子名称: | 1-[(3R,4R)-3-(hydroxymethyl)-4-phenylpyrrolidin-1-yl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 3C | | 著者 | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-24 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Crystallographic Fragment Screen of Coxsackievirus A16 2A Protease identifies new opportunities for the development of broad-spectrum anti-enterovirals.
Biorxiv, 2024
|
|
2BXQ
 
 | | Human serum albumin complexed with myristate, phenylbutazone and indomethacin | | 分子名称: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, INDOMETHACIN, MYRISTIC ACID, ... | | 著者 | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | 登録日 | 2005-07-26 | | 公開日 | 2005-09-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
4RW5
 
 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | 分子名称: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | 著者 | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | 登録日 | 2014-12-01 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
1JP3
 
 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | 分子名称: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | 著者 | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | 登録日 | 2001-07-31 | | 公開日 | 2001-08-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
6FDC
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-32a | | 分子名称: | (2~{R})-1-[3-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, (2~{S})-1-[5-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]pyrazol-1-yl]-3-morpholin-4-yl-propan-2-ol, 1,2-ETHANEDIOL, ... | | 著者 | Prosdocimi, T, Donini, S, Parisini, E. | | 登録日 | 2017-12-22 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
4DY6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 2'-phosphate bis(adenosine)-5'-diphosphate | | 分子名称: | CITRIC ACID, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, inorganic polyphosphate/ATP-NAD kinase 1 | | 著者 | Poncet-Montange, G, Assairi, L, Arold, S, Pochet, S, Labesse, G. | | 登録日 | 2012-02-28 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
2V0N
 
 | | ACTIVATED RESPONSE REGULATOR PLED IN COMPLEX WITH C-DIGMP AND GTP- ALPHA-S | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), BERYLLIUM TRIFLUORIDE ION, CHLORIDE ION, ... | | 著者 | Wassmann, P, Schirmer, T. | | 登録日 | 2007-05-15 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structure of Bef3--Modified Response Regulator Pled: Implications for Diguanylate Cyclase Activation, Catalysis, and Feedback Inhibition
Structure, 15, 2007
|
|
3UJ4
 
 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | 著者 | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | 登録日 | 2011-11-07 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
4S18
 
 | |
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | 分子名称: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | 著者 | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | 登録日 | 2020-02-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1OYV
 
 | | Crystal structure of tomato inhibitor-II in a ternary complex with subtilisin Carlsberg | | 分子名称: | CALCIUM ION, Subtilisin Carlsberg, Wound-induced proteinase inhibitor-II | | 著者 | Barrette-Ng, I.H, Ng, K.K, Cherney, M.M, Pearce, G, Ryan, C.A, James, M.N. | | 登録日 | 2003-04-07 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg
J.Biol.Chem., 278, 2003
|
|
5V71
 
 | | KRAS G12C in bound to quinazoline based switch II pocket (SWIIP) binder | | 分子名称: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Westover, K, Lu, J. | | 登録日 | 2017-03-17 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.228 Å) | | 主引用文献 | KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry.
Structure, 25, 2017
|
|
4CFK
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH LY294002 | | 分子名称: | 1,2-ETHANEDIOL, 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, BRD4 PROTEIN, ... | | 著者 | Chung, C, Dittmann, A, Drewes, G. | | 登録日 | 2013-11-18 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Commonly Used Pi3-Kinase Probe Ly294002 is an Inhibitor of Bet Bromodomains.
Acs Chem.Biol., 9, 2014
|
|
4E2T
 
 | | Crystal Structures of RadA intein from Pyrococcus horikoshii | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
3R67
 
 | |
5VC3
 
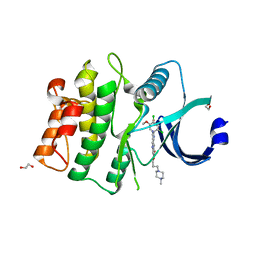 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH BOSUTINIB | | 分子名称: | 1,2-ETHANEDIOL, 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, CHLORIDE ION, ... | | 著者 | Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2017-03-30 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5RF0
 
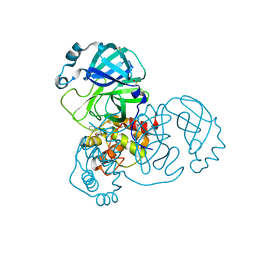 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0073 | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, [1-(pyridin-2-yl)cyclopentyl]methanol | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5UMC
 
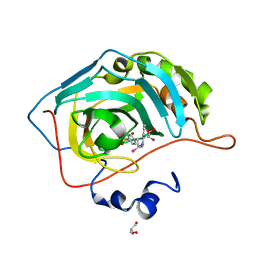 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | 分子名称: | Carbonic anhydrase 2, GLYCEROL, UNKNOWN LIGAND, ... | | 著者 | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | 登録日 | 2017-01-26 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
