5OY3
 
 | | The structural basis of the histone demethylase KDM6B histone 3 lysine 27 specificity | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | 著者 | Jones, S.E, Olsen, L, Gajhede, M. | | 登録日 | 2017-09-07 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.136 Å) | | 主引用文献 | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
3EQP
 
 | | Crystal Structure of Ack1 with compound T95 | | 分子名称: | Activated CDC42 kinase 1, CHLORIDE ION, N-(2,6-dimethylphenyl)-4-(2-ethoxyphenoxy)-2-({4-[4-(2-hydroxyethyl)piperazin-1-yl]phenyl}amino)pyrimidine-5-carboxamide | | 著者 | Liu, J, Wang, Z, Walker, N.P.C. | | 登録日 | 2008-10-01 | | 公開日 | 2008-12-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3EB1
 
 | | Crystal structure PTP1B complex with small molecule inhibitor LZP-25 | | 分子名称: | 4-[3-(dibenzylamino)phenyl]-2,4-dioxobutanoic acid, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Liu, S, Zheng, L.-F, Wu, L, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q, Zhang, Z.-Y. | | 登録日 | 2008-08-26 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
1Q0Q
 
 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | 分子名称: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | 登録日 | 2003-07-17 | | 公開日 | 2004-07-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
4XI8
 
 | |
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | 分子名称: | De novo designed diiron protein, ZINC ION | | 著者 | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | 登録日 | 2006-08-08 | | 公開日 | 2007-07-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
2X1N
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | 分子名称: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | 著者 | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | 登録日 | 2009-12-31 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
1O97
 
 | | Structure of electron transferring flavoprotein from Methylophilus methylotrophus, recognition loop removed by limited proteolysis | | 分子名称: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | 著者 | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | 登録日 | 2002-12-11 | | 公開日 | 2003-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
6EET
 
 | | Crystal structure of mouse Protocadherin-15 EC9-MAD12 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Narui, Y, Sotomayor, M. | | 登録日 | 2018-08-15 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5OP9
 
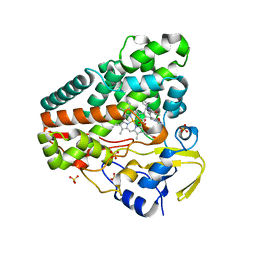 | | The crystal structure of P450 CYP121 in complex with lead compound 7e | | 分子名称: | 4-(imidazol-1-ylmethyl)-3-(4-methoxyphenyl)-1-phenyl-pyrazole, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Levy, C.W. | | 登録日 | 2017-08-09 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.455 Å) | | 主引用文献 | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
5OPA
 
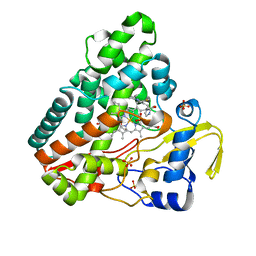 | | The crystal structure of P450 CYP121 in complex with lead compound 7b | | 分子名称: | 3-(4-fluorophenyl)-4-(imidazol-1-ylmethyl)-1-phenyl-pyrazole, DI(HYDROXYETHYL)ETHER, Mycocyclosin synthase, ... | | 著者 | Levy, C.W. | | 登録日 | 2017-08-09 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.345 Å) | | 主引用文献 | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
5O4L
 
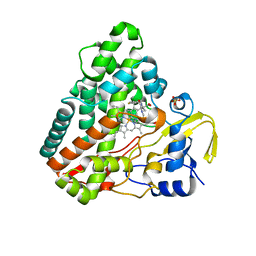 | | Crystal structure of P450 CYP121 in complex with compound 6a. | | 分子名称: | 1-[(4-fluorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Levy, C.W. | | 登録日 | 2017-05-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
1TSK
 
 | |
3WI3
 
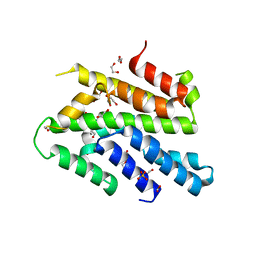 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | 分子名称: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | 著者 | Itou, H, Araki, H, Shirakihara, Y. | | 登録日 | 2013-09-05 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
5NFB
 
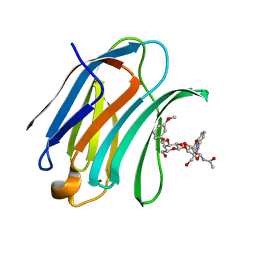 | | Structure of Galectin-3 CRD in complex with compound 4 | | 分子名称: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-4-[(3-methoxyphenyl)methoxy]-3,5-bis(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | 著者 | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | 登録日 | 2017-03-13 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5NF9
 
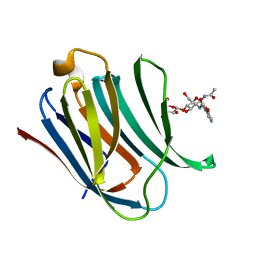 | | Structure of Galectin-3 CRD in complex with compound 2 | | 分子名称: | Galectin-3, ~{N}-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-acetamido-6-(hydroxymethyl)-5-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4-oxidanyl-oxan-3-yl]-3-methoxy-benzamide | | 著者 | Ronin, C, Atmanene, C, Gautier, F.M, Djedaini Pilard, F, Teletchea, S, Ciesielski, F, Vivat Hannah, V, Grandjean, C. | | 登録日 | 2017-03-13 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5O4K
 
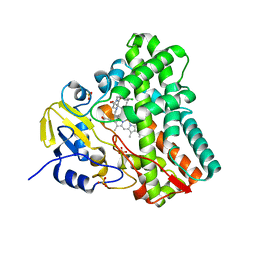 | | Crystal structure of P450 CYP121 in complex with compound 6b. | | 分子名称: | 1-[(4-chlorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Levy, C.W. | | 登録日 | 2017-05-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
1O96
 
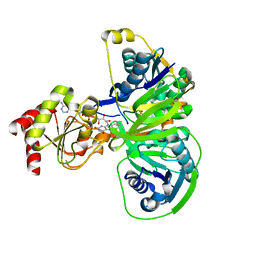 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | 分子名称: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | 著者 | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | 登録日 | 2002-12-11 | | 公開日 | 2003-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
2YN8
 
 | |
4ZB7
 
 | |
4ZBD
 
 | |
6FQ3
 
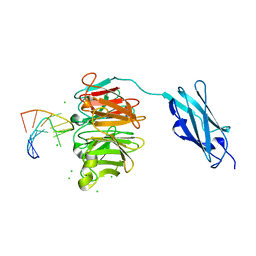 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | 著者 | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | 登録日 | 2018-02-13 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
4Z8B
 
 | | crystal structure of a DGL mutant - H51G H131N | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, GLYCEROL, ... | | 著者 | Zamora-Caballero, S, Perez, A, Sanz, L, Bravo, J, Calvete, J.J. | | 登録日 | 2015-04-08 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Quaternary structure of Dioclea grandiflora lectin assessed by equilibrium sedimentation and crystallographic analysis of recombinant mutants.
Febs Lett., 589, 2015
|
|
4ZB6
 
 | |
3MB8
 
 | | Crystal structure of purine nucleoside phosphorylase from toxoplasma gondii in complex with immucillin-H | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Ho, M, Almo, S.C, Schramm, V.L. | | 登録日 | 2010-03-25 | | 公開日 | 2011-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Eukaryot Cell, 13, 2014
|
|
