3UC8
 
 | | Trp-cage cyclo-TC1 - tetragonal crystal form | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, cyclo-TC1 | | 著者 | Scian, M, Le Trong, I, Stenkamp, R.E, Andersen, N.H. | | 登録日 | 2011-10-26 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UCJ
 
 | | Coccomyxa beta-carbonic anhydrase in complex with acetazolamide | | 分子名称: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CHLORIDE ION, Carbonic anhydrase, ... | | 著者 | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | 登録日 | 2011-10-27 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-12-25 | | 公開日 | 2014-03-05 | | 最終更新日 | 2015-02-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6PXD
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | 著者 | Du, J, Lu, W, Gouaux, E. | | 登録日 | 2019-07-25 | | 公開日 | 2021-02-10 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6Z4C
 
 | | The structure of the N-terminal domain of RssB from E. coli | | 分子名称: | Regulator of RpoS | | 著者 | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | 登録日 | 2020-05-25 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | 分子名称: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | 著者 | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | 登録日 | 2014-01-09 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | 分子名称: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
6DKF
 
 | |
3UF0
 
 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-10-31 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
5D9R
 
 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | 分子名称: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | 著者 | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
3UQP
 
 | | Crystal structure of Bace1 with its inhibitor | | 分子名称: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | 著者 | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | 登録日 | 2011-11-21 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
3USJ
 
 | |
3UMK
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with copper | | 分子名称: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | 著者 | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | 登録日 | 2011-11-13 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
4O74
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with BI 2536 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4, ... | | 著者 | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | 登録日 | 2013-12-24 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | 著者 | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | 登録日 | 2011-10-27 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3USK
 
 | |
4O9A
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | 分子名称: | Acetyl-CoA acetyltransferase | | 著者 | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | 登録日 | 2014-01-02 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
3UG9
 
 | | Crystal Structure of the Closed State of Channelrhodopsin | | 分子名称: | Archaeal-type opsin 1, Archaeal-type opsin 2, OLEIC ACID, ... | | 著者 | Kato, H.E, Ishitani, R, Nureki, O. | | 登録日 | 2011-11-02 | | 公開日 | 2012-01-25 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the channelrhodopsin light-gated cation channel
Nature, 482, 2012
|
|
1MEK
 
 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 40 STRUCTURES | | 分子名称: | PROTEIN DISULFIDE ISOMERASE | | 著者 | Kemmink, J, Darby, N.J, Dijkstra, K, Nilges, M, Creighton, T.E. | | 登録日 | 1996-04-16 | | 公開日 | 1997-04-21 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy.
Biochemistry, 35, 1996
|
|
6DLU
 
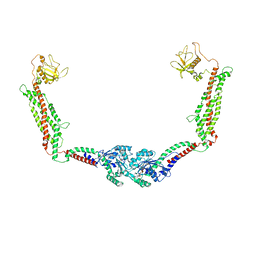 | | Cryo-EM of the GMPPCP-bound human dynamin-1 polymer assembled on the membrane in the constricted state | | 分子名称: | Dynamin-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | 著者 | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | 登録日 | 2018-06-02 | | 公開日 | 2018-08-01 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
6DMR
 
 | |
4OVL
 
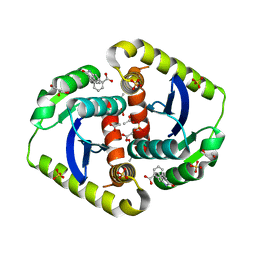 | |
3UOD
 
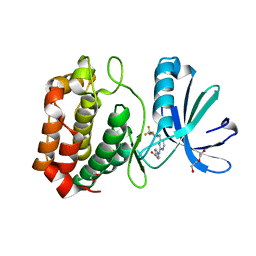 | | Aurora A in complex with RPM1693 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-{[2-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzoic acid, Aurora kinase A, ... | | 著者 | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2011-11-16 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5002 Å) | | 主引用文献 | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
5D1Y
 
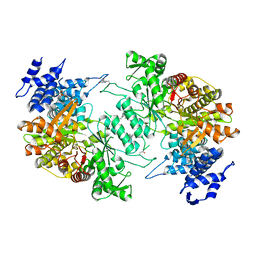 | | Low resolution crystal structure of human ribonucleotide reductase alpha6 hexamer in complex with dATP | | 分子名称: | Ribonucleoside-diphosphate reductase large subunit | | 著者 | Ando, N, Li, H, Brignole, E.J, Thompson, S, McLaughlin, M.I, Page, J, Asturias, F, Stubbe, J, Drennan, C.L. | | 登録日 | 2015-08-04 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (9.005 Å) | | 主引用文献 | Allosteric Inhibition of Human Ribonucleotide Reductase by dATP Entails the Stabilization of a Hexamer.
Biochemistry, 55, 2016
|
|
4ODQ
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with S3 peptide | | 分子名称: | 30S ribosomal protein S3, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Quistgaard, E.M, Low, C, Nordlund, P. | | 登録日 | 2014-01-10 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
