8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-08-01 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | 登録日 | 2022-08-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | 分子名称: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | 登録日 | 2019-09-20 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
2B5L
 
 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | 分子名称: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | 著者 | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | 登録日 | 2005-09-28 | | 公開日 | 2006-02-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
8KG8
 
 | | Yeast replisome in state II | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.23 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8W7S
 
 | | Yeast replisome in state IV | | 分子名称: | Cell division control protein 45, DNA (71-mer), DNA polymerase alpha-binding protein, ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D. | | 登録日 | 2023-08-31 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (7.39 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG6
 
 | | Yeast replisome in state I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG9
 
 | | Yeast replisome in state III | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-17 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.52 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | 分子名称: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | 著者 | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | 登録日 | 2022-07-19 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | 分子名称: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | 著者 | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | 登録日 | 2022-07-19 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8W7M
 
 | | Yeast replisome in state V | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (71-mer), ... | | 著者 | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | 登録日 | 2023-08-30 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
2B5M
 
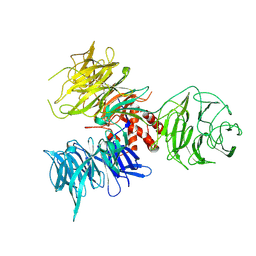 | | Crystal Structure of DDB1 | | 分子名称: | damage-specific DNA binding protein 1 | | 著者 | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | 登録日 | 2005-09-28 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
1MA7
 
 | |
6FE8
 
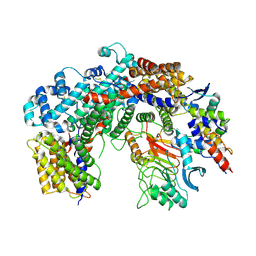 | | Cryo-EM structure of the core Centromere Binding Factor 3 complex | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | 著者 | Zhang, W.J, Lukoynova, N, Miah, S, Vaughan, C.K. | | 登録日 | 2017-12-30 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
1BIA
 
 | |
6GSA
 
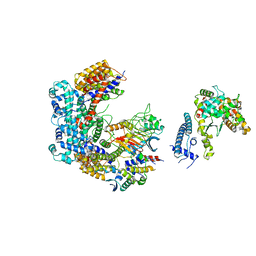 | | Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Zhang, W.J, Lukoynova, N, Vaughan, C.K. | | 登録日 | 2018-06-13 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10.
Cell Rep, 24, 2018
|
|
1BIB
 
 | |
3CRX
 
 | |
7ONI
 
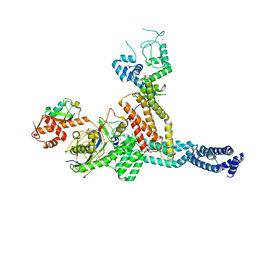 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | 分子名称: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | 著者 | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | 登録日 | 2021-05-25 | | 公開日 | 2021-09-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
7MWD
 
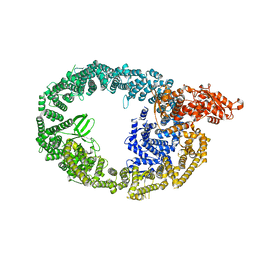 | | HUWE1 in map with focus on HECT | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Hunkeler, M, Fischer, E.S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MWF
 
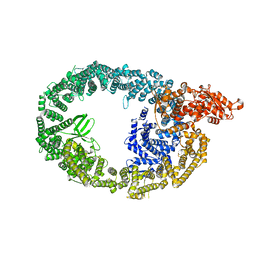 | |
7MWE
 
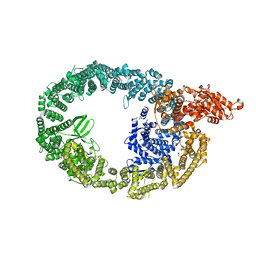 | | HUWE1 in map with focus on WWE | | 分子名称: | E3 ubiquitin-protein ligase HUWE1 | | 著者 | Hunkeler, M, Fischer, E.S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
8BJA
 
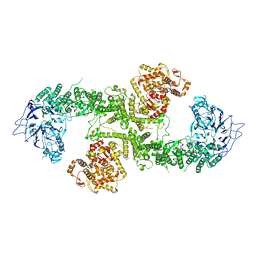 | |
7XZZ
 
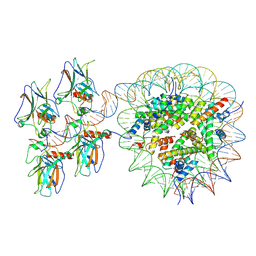 | | Cryo-EM structure of the nucleosome in complex with p53 | | 分子名称: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | 著者 | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-06-03 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.07 Å) | | 主引用文献 | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
8B5W
 
 | | Crystal structure of the E3 module from UBR4 | | 分子名称: | 1,2-ETHANEDIOL, ZINC ION, cDNA FLJ12511 fis, ... | | 著者 | Virdee, S, Mabbitt, P.D, Barnsby-Greer, L. | | 登録日 | 2022-09-24 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4.
Nat.Struct.Mol.Biol., 31, 2024
|
|
