2C62
 
 | |
5XE7
 
 | |
6YWN
 
 | | CutA in complex with CMPCPP | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CALCIUM ION, CutA | | 著者 | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | 登録日 | 2020-04-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2020-09-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6NZ2
 
 | |
6YWP
 
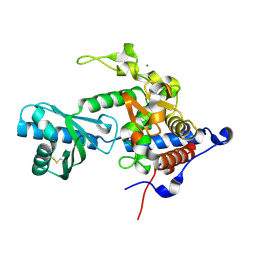 | | Structure of apo-CutA | | 分子名称: | CutA, MAGNESIUM ION | | 著者 | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | 登録日 | 2020-04-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2020-09-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6BCD
 
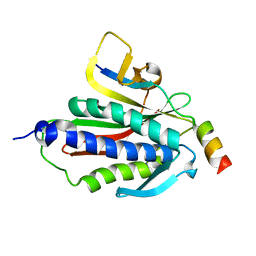 | | Crystal structure of Rev7-K44A/R124A/A135D in complex with Rev3-RBM2 (residues 1988-2014) | | 分子名称: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7M09
 
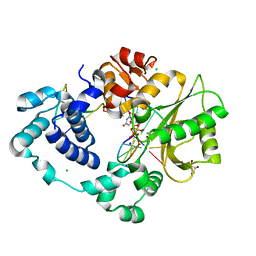 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with blunt-ended DSB substrate and incoming dUMPNPP | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0D
 
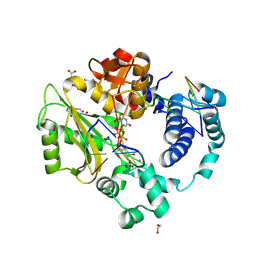 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M0B
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound mismatched DSB and incoming dUMPNPP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7M07
 
 | | Pre-catalytic ternary complex of DNA Polymerase Lambda with bound 1-nt gapped SSB substrate and incoming dUMPNPP | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
6BC8
 
 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | 分子名称: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | 著者 | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | 登録日 | 2017-10-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6XEO
 
 | | Structure of Mfd bound to dsDNA | | 分子名称: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | 著者 | Brugger, C, Deaconescu, A. | | 登録日 | 2020-06-12 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
4P9A
 
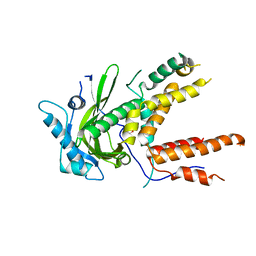 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | 分子名称: | GLYCEROL, Polymerase PA | | 著者 | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2014-04-02 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1999 Å) | | 主引用文献 | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
6BI7
 
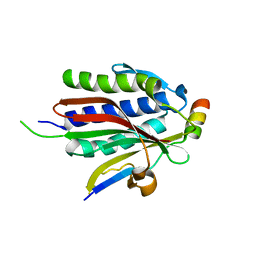 | | Crystal structure of Rev7-WT/Rev3 as a monomer under high-salt conditions | | 分子名称: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Rizzo, A.A, Korzhnev, D.M, Hao, B, Li, Y. | | 登録日 | 2017-11-01 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2HHX
 
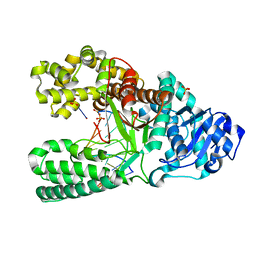 | | O6-methyl-guanine in the polymerase template preinsertion site | | 分子名称: | 5'-D(*CP*AP*TP*(6OG)P*CP*GP*AP*GP*TP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*G)-3', DNA Polymerase I, ... | | 著者 | Warren, J.J, Forsberg, L.J, Beese, L.S. | | 登録日 | 2006-06-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6FS6
 
 | |
2HHU
 
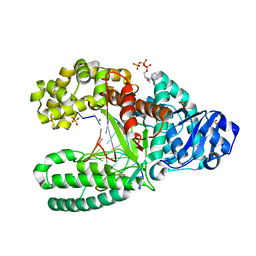 | | C:O6-methyl-guanine in the polymerase postinsertion site (-1 basepair position) | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*CP*C)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*GP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', ... | | 著者 | Warren, J.J, Forsberg, L.J, Beese, L.S. | | 登録日 | 2006-06-28 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6FS7
 
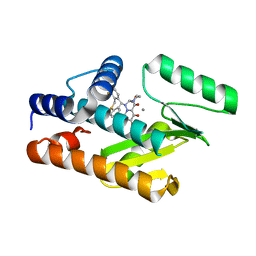 | |
4OR6
 
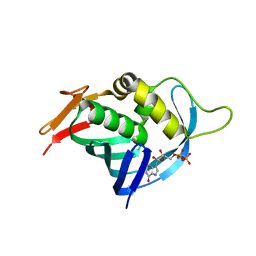 | |
2IQH
 
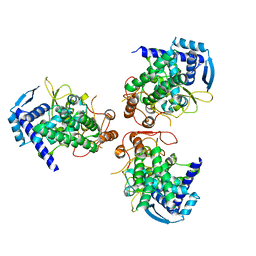 | |
4Q46
 
 | |
6Z3U
 
 | | Structure of the CAK complex form Chaetomium thermophilum | | 分子名称: | CHLORIDE ION, CYCLIN domain-containing protein, Protein kinase domain-containing protein, ... | | 著者 | Peissert, S, Kuper, J, Kisker, C. | | 登録日 | 2020-05-22 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PFJ
 
 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | 著者 | Schumacher, M.A. | | 登録日 | 2019-06-21 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6USR
 
 | |
6ONO
 
 | | Complex structure of WhiB1 and region 4 of SigA in C2221 space group | | 分子名称: | DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, RNA polymerase sigma factor SigA, ... | | 著者 | Wan, T, Li, S.R, Beltran, D.G, Schacht, A, Becker, D.C, Zhang, L.M. | | 登録日 | 2019-04-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2020-01-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis of non-canonical transcriptional regulation by the sigma A-bound iron-sulfur protein WhiB1 in M. tuberculosis.
Nucleic Acids Res., 48, 2020
|
|
