7LUD
 
 | | Crystal structure of Fab ADI-14442 | | 分子名称: | ADI-14442 Fab Heavy chain, ADI-14442 Fab Light chain, SULFATE ION | | 著者 | Rush, S.A, McLellan, J.S. | | 登録日 | 2021-02-22 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses.
Immunity, 54, 2021
|
|
6C2I
 
 | |
6C2R
 
 | | Aurora A ligand complex | | 分子名称: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | 著者 | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | 登録日 | 2018-01-08 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6CQO
 
 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae (SeMet Labeled), Rim1 (Form2) | | 分子名称: | Single-stranded DNA-binding protein RIM1, mitochondrial | | 著者 | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | 登録日 | 2018-03-15 | | 公開日 | 2018-05-30 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | 分子名称: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MKM
 
 | |
7MKL
 
 | |
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | 著者 | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2021-05-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
2PU4
 
 | |
7MS7
 
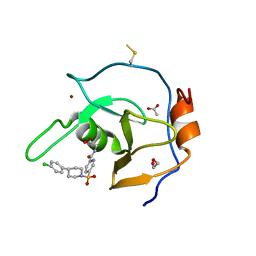 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | 著者 | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | 登録日 | 2021-05-10 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | 分子名称: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | 著者 | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | 登録日 | 2021-05-10 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | 分子名称: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | 著者 | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | 登録日 | 2021-05-10 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7K99
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | 分子名称: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2020-09-28 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7JO1
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butyramide | | 分子名称: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)butanamide, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JOB
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | 分子名称: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-06 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.381 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
6BVU
 
 | | SFTI-HFRW-1 | | 分子名称: | Trypsin inhibitor 1 HFRW-1 | | 著者 | Schroeder, C.I. | | 登録日 | 2017-12-13 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Development of Novel Melanocortin Receptor Agonists Based on the Cyclic Peptide Framework of Sunflower Trypsin Inhibitor-1.
J.Med.Chem., 61, 2018
|
|
7K3T
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | 著者 | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-13 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-08-19 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
2OTY
 
 | | 1,2-dichlorobenzene in complex with T4 Lysozyme L99A | | 分子名称: | 1,2-DICHLOROBENZENE, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Graves, A.P, Shoichet, B.K. | | 登録日 | 2007-02-09 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Predicting absolute ligand binding free energies to a simple model site.
J.Mol.Biol., 371, 2007
|
|
6B1F
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by soaking | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, 1,2-ETHANEDIOL, CITRIC ACID, ... | | 著者 | van den Akker, F, Nguyen, N.Q. | | 登録日 | 2017-09-18 | | 公開日 | 2018-08-01 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1H
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by co-crystallization | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | 著者 | van den Akker, F, Nhuyen, N.Q. | | 登録日 | 2017-09-18 | | 公開日 | 2018-08-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
6B1R
 
 | |
7K88
 
 | | Crystal structure of bovine RPE65 in complex with hexaethylene glycol monooctyl ether | | 分子名称: | 3,6,9,12,15,18-hexaoxahexacosan-1-ol, FE (II) ION, Retinoid isomerohydrolase, ... | | 著者 | Kiser, P.D. | | 登録日 | 2020-09-26 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy.
J.Med.Chem., 64, 2021
|
|
7K8G
 
 | |
7K89
 
 | |
