7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | 分子名称: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-12-12 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AQI
 
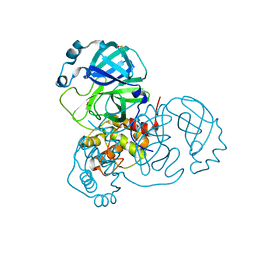 | | Structure of SARS-CoV-2 Main Protease bound to Ifenprodil | | 分子名称: | 3C-like proteinase, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, DIMETHYL SULFOXIDE, ... | | 著者 | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-21 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR5
 
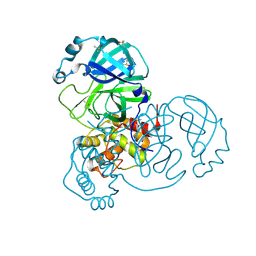 | | Structure of apo SARS-CoV-2 Main Protease with small beta angle, space group C2. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1G49
 
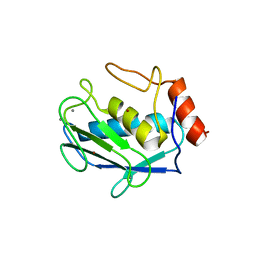 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | 分子名称: | (1N)-4-N-BUTOXYPHENYLSULFONYL-(2R)-N-HYDROXYCARBOXAMIDO-(4S)-METHANESULFONYLAMINO-PYRROLIDINE, CALCIUM ION, MATRIX METALLOPROTEINASE 3, ... | | 著者 | Natchus, M.G, Bookland, R.G, De, B, Almstead, N.G, Pikul, S. | | 登録日 | 2000-10-26 | | 公開日 | 2001-10-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of new hydroxamate matrix metalloproteinase inhibitors derived from functionalized 4-aminoprolines.
J.Med.Chem., 43, 2000
|
|
7ASS
 
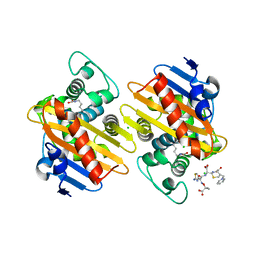 | |
7AF0
 
 | | Structure of SARS-CoV-2 Main Protease bound to Ipidacrine. | | 分子名称: | 2,3,5,6,7,8-hexahydro-1~{H}-cyclopenta[b]quinolin-9-amine, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-18 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6WOJ
 
 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | 登録日 | 2020-04-24 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
7APH
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1FU7
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | 分子名称: | GLYCOGEN PHOSPHORYLASE, N-(methoxycarbonyl)-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | 登録日 | 2000-09-14 | | 公開日 | 2000-10-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6W9T
 
 | |
6NV9
 
 | | BACE1 in complex with a macrocyclic inhibitor | | 分子名称: | (3S)-3-hydroxy-N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, Beta-secretase 1, SULFATE ION | | 著者 | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | 登録日 | 2019-02-04 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
6NW3
 
 | | BACE1 in complex with a macrocyclic inhibitor | | 分子名称: | Beta-secretase 1, N-(2-methylpropyl)-N~2~-{[(4S)-17-[(methylsulfonyl)(propyl)amino]-2-oxo-3-azatricyclo[13.3.1.1~6,10~]icosa-1(19),6(20),7,9,15,17-hexaen-4-yl]methyl}-L-norleucinamide, SULFATE ION | | 著者 | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | 登録日 | 2019-02-05 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.352 Å) | | 主引用文献 | Development of an Efficient Enzyme Production and Structure-Based Discovery Platform for BACE1 Inhibitors.
Biochemistry, 58, 2019
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | 分子名称: | RNA (171-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
1FU8
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | 分子名称: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | 登録日 | 2000-09-14 | | 公開日 | 2000-10-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6WLJ
 
 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | 分子名称: | RNA (130-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (9.6 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
1FWY
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | 著者 | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | 登録日 | 2000-09-25 | | 公開日 | 2000-10-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
7CBI
 
 | | Crystal structure of threonyl-tRNA synthetase (ThrRS) from Salmonella enterica in complex with an inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl (2~{S},3~{R})-2-azanyl-3-oxidanyl-butanoate, GLYCEROL, ... | | 著者 | Guo, J, Chen, B, Zhou, H. | | 登録日 | 2020-06-12 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structure-guided optimization and mechanistic study of a class of quinazolinone-threonine hybrids as antibacterial ThrRS inhibitors.
Eur.J.Med.Chem., 207, 2020
|
|
6WLN
 
 | | hc16 ligase product models, 10.0 Angstrom resolution | | 分子名称: | RNA (349-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
1FTY
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | 分子名称: | 8,9,10-TRIHYDROXY-7-HYDROXYMETHYL-3-METHYL-6-OXA-1,3-DIAZA-SPIRO[4.5]DECANE-2,4-DIONE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | 登録日 | 2000-09-13 | | 公開日 | 2000-10-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6WLQ
 
 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | 分子名称: | RNA (119-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLO
 
 | | hc16 ligase models, 11.0 Angstrom resolution | | 分子名称: | RNA (338-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WEA
 
 | |
1GG8
 
 | | DESIGN OF INHIBITORS OF GLYCOGEN PHOSPHORYLASE: A STUDY OF ALPHA-AND BETA-C-GLUCOSIDES AND 1-THIO-BETA-D-GLUCOSE COMPOUNDS | | 分子名称: | ALPHA-D-GLUCOPYRANOSYL-2-CARBOXYLIC ACID AMIDE, INOSINIC ACID, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | 著者 | Watson, K.A, Mitchell, E.P, Johnson, L.N, Son, J.C, Bichard, C.J, Orchard, M.G, Fleet, G.W, Oikonomakos, N.G, Leonidas, D.D, Kontou, M, Papageorgiou, A.C. | | 登録日 | 2000-07-30 | | 公開日 | 2000-08-23 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Design of inhibitors of glycogen phosphorylase: a study of alpha- and beta-C-glucosides and 1-thio-beta-D-glucose compounds.
Biochemistry, 33, 1994
|
|
6WLU
 
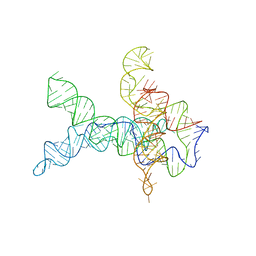 | | V. cholerae glycine riboswitch with glycine models, 5.7 Angstrom resolution | | 分子名称: | RNA (231-MER) | | 著者 | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | 登録日 | 2020-04-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
7C0P
 
 | |
