3L3O
 
 | |
1I83
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH N1,N14-BIS((S-METHYL)ISOTHIOUREIDO)TETRADECANE (H4B FREE) | | 分子名称: | ACETATE ION, GLYCEROL, N1,N14-BIS((S-METHYL)ISOTHIOUREIDO)TETRADECANE, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Babu, B.R, Griffith, O.W, Masters, B.S.S, Poulos, T.L. | | 登録日 | 2001-03-12 | | 公開日 | 2001-08-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
4CFN
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(cyclohexylmethoxy)-8-(trifluoromethyl)-9H-purin-2-amine, CYCLIN-A2, ... | | 著者 | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | 登録日 | 2013-11-19 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
1I2N
 
 | |
6MUL
 
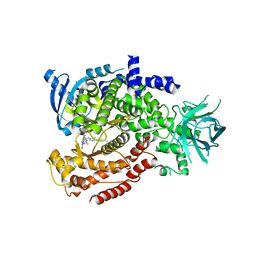 | | Murine PI3K delta kinsae domain - cpd 1 | | 分子名称: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Fischmann, T.O. | | 登録日 | 2018-10-23 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
1N1O
 
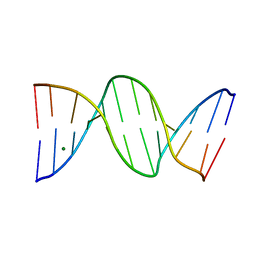 | | Crystal Structure of a B-form DNA Duplex Containing (L)-alpha-threofuranosyl (3'-2') Nucleosides: A Four-Carbon Sugar is Easily Accommodated into the Backbone of DNA | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*(TFT)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | 著者 | Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | 登録日 | 2002-10-18 | | 公開日 | 2002-11-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal Structure of a B-Form DNA Duplex Containing (L)-alpha-Threofuranosyl (3'-->2') Nucleosides: A
Four-Carbon Sugar Is Easily Accommodated into the Backbone of DNA
J.Am.Chem.Soc., 124, 2002
|
|
6BWL
 
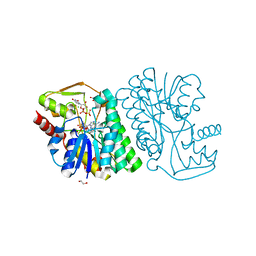 | | X-ray structure of Pal from Bacillus thuringiensis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | 登録日 | 2017-12-15 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
5SZO
 
 | | Protocadherin Gamma B7 extracellular cadherin domains 1-4 P41212 crystal form | | 分子名称: | CALCIUM ION, Protocadherin Gamma B7, alpha-D-mannopyranose, ... | | 著者 | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | 登録日 | 2016-08-14 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.612 Å) | | 主引用文献 | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5UNM
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, substrate free form with flexible loop | | 分子名称: | ATP-utilizing enzyme of the PP-loopsuperfamily, PHOSPHATE ION | | 著者 | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | 登録日 | 2017-01-31 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
1K8L
 
 | | XBY6: An analog of CK14 containing 6 dithiophosphate groups | | 分子名称: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | 著者 | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | 登録日 | 2001-10-24 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
6BTM
 
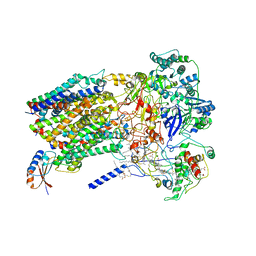 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | 著者 | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | 登録日 | 2017-12-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
4YMR
 
 | |
1GJF
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | 分子名称: | IGFBP-1 antagonist, PENTANE | | 著者 | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | 登録日 | 2001-05-11 | | 公開日 | 2001-05-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1K8J
 
 | | NMR STRUCTURE OF THE CK14 DNA DUPLEX: A PORTION OF THE KNOWN NF-kB SEQUENCE CK1 | | 分子名称: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | 著者 | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | 登録日 | 2001-10-24 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
1K8N
 
 | | NMR structure of the XBY2 DNA duplex, an analog of CK14 containing phosphorodithioate groups at C22 and C24 | | 分子名称: | FIRST STRAND OF CK14 DNA DUPLEX, SECOND STRAND OF CK14 DNA DUPLEX | | 著者 | Volk, D.E, Yang, X, Fennewald, S.M, King, D.J, Bassett, S.E, Venkitachalam, S, Herzog, N, Luxon, B.A, Gorenstein, D.G. | | 登録日 | 2001-10-24 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-kappaB
Bioorg.Chem., 30, 2002
|
|
5TQY
 
 | | CryoEM reconstruction of human IKK1, closed conformation 3 | | 分子名称: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | 著者 | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | 登録日 | 2016-10-24 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
2G5K
 
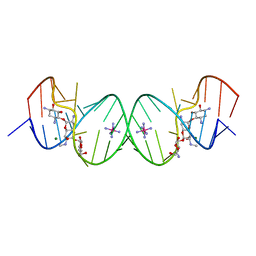 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with Apramycin | | 分子名称: | 5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3', APRAMYCIN, COBALT HEXAMMINE(III), ... | | 著者 | Kondo, J, Francois, B, Urzhumtsev, A, Westhof, E. | | 登録日 | 2006-02-23 | | 公開日 | 2006-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site Complexed with Apramycin
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
8HRY
 
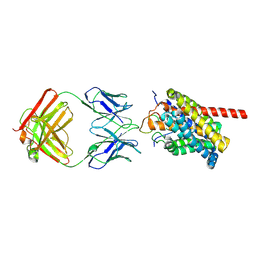 | | Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex | | 分子名称: | Fab heavy chain from antibody IgG clone number YN9016, Fab light chain from antibody IgG clone number YN9016, Large S protein (Fragment), ... | | 著者 | Asami, J, Shimizu, T, Ohto, U. | | 登録日 | 2022-12-16 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HFI
 
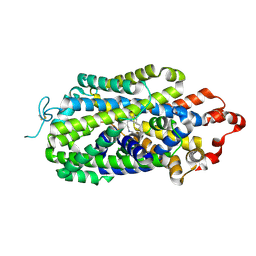 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant desipramine in an inward-open state at resolution of 2.5 angstrom. | | 分子名称: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | 著者 | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | 登録日 | 2022-11-10 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
1JLB
 
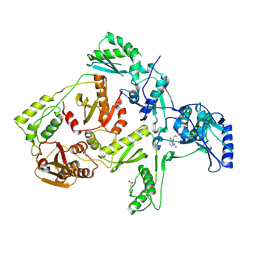 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | 著者 | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | 登録日 | 2001-07-16 | | 公開日 | 2001-10-03 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4LJP
 
 | | Structure of an active ligase (HOIP-H889A)/ubiquitin transfer complex | | 分子名称: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-C, ZINC ION | | 著者 | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | 登録日 | 2013-07-05 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
4D6F
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | 分子名称: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | 登録日 | 2014-11-11 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6I
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | 分子名称: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | 登録日 | 2014-11-11 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5GVW
 
 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | 分子名称: | Glycosyl transferase family 8, MANGANESE (II) ION | | 著者 | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | 登録日 | 2016-09-07 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
