7DCU
 
 | |
7DCT
 
 | |
7R1Y
 
 | |
7DCI
 
 | |
6FT3
 
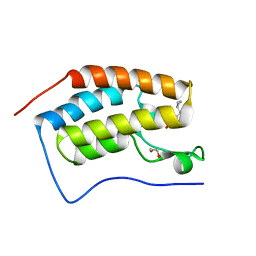 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6H4Z
 
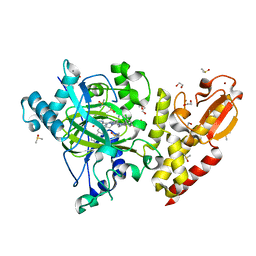 | | Crystal structure of human KDM5B in complex with compound 16a | | 分子名称: | 1,2-ETHANEDIOL, 8-[4-[2-[4-(3-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | 著者 | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | 登録日 | 2018-07-23 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
7DCS
 
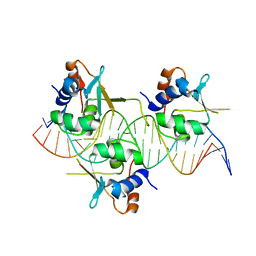 | |
6H50
 
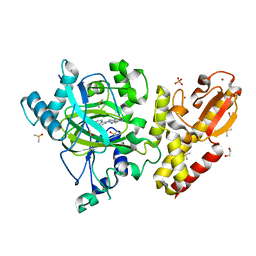 | | Crystal structure of human KDM5B in complex with compound 34a | | 分子名称: | 1,2-ETHANEDIOL, 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | 著者 | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | 登録日 | 2018-07-23 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6FY5
 
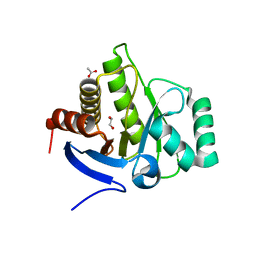 | |
6CBI
 
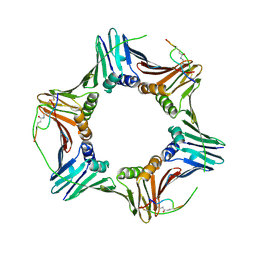 | | PCNA in complex with inhibitor | | 分子名称: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | 著者 | Bruning, J.B, Wegener, K.L. | | 登録日 | 2018-02-03 | | 公開日 | 2018-07-04 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
3ZMS
 
 | | LSD1-CoREST in complex with INSM1 peptide | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, INSULINOMA-ASSOCIATED PROTEIN 1, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, ... | | 著者 | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | 登録日 | 2013-02-12 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6ZN6
 
 | | Protein polybromo-1 (PB1 BD2) Bound To MW278 | | 分子名称: | 1,2-ETHANEDIOL, 2-[6-azanyl-5-[(3~{R})-3-phenoxypiperidin-1-yl]pyridazin-3-yl]phenol, Protein polybromo-1 | | 著者 | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | 登録日 | 2020-07-06 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZNO
 
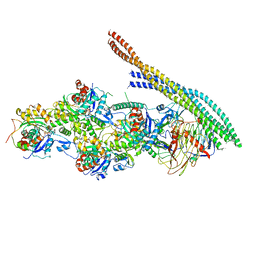 | | The pointed end complex of dynactin with the p150 projection docked | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | 著者 | Lau, C.K, Lacey, S.E, Carter, A.P. | | 登録日 | 2020-07-06 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
7A1F
 
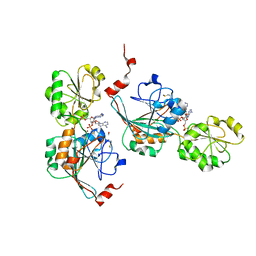 | | Crystal structure of human 5' exonuclease Appollo in complex with 5'dAMP | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5' exonuclease Apollo, FE (III) ION, ... | | 著者 | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-08-12 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
2KXN
 
 | | NMR structure of human Tra2beta1 RRM in complex with AAGAAC RNA | | 分子名称: | 5'-R(*AP*AP*GP*AP*AP*C)-3', Transformer-2 protein homolog beta | | 著者 | Clery, A, Jayne, S, Benderska, N, Dominguez, C, Stamm, S, Allain, F.H.-T. | | 登録日 | 2010-05-10 | | 公開日 | 2011-03-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Nat.Struct.Mol.Biol., 18, 2011
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | 分子名称: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-02-20 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6ZOK
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3UJF
 
 | | Asymmetric complex of human neuron specific enolase-4-PGA/PEP | | 分子名称: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | 著者 | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | 登録日 | 2011-11-07 | | 公開日 | 2012-08-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
6GZH
 
 | | Crystal Structure of Human CDK9/cyclinT1 with A86 | | 分子名称: | Cyclin-T1, Cyclin-dependent kinase 9, GLYCEROL, ... | | 著者 | Ben-neriah, Y, Venkatachalam, A, Minzel, W, Fink, A, Snir-Alkalay, I, Vacca, J. | | 登録日 | 2018-07-04 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Small Molecules Co-targeting CKI alpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models.
Cell, 175, 2018
|
|
6ZO4
 
 | | The pointed end complex of dynactin bound to BICD2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | 著者 | Lau, C.K, Lacey, S.E, Carter, A.P. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-02 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
8JDL
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
6C83
 
 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | 分子名称: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | 登録日 | 2018-01-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
