8QDR
 
 | | Vitis vinifera dimeric 13S-lipoxygenase LOXA in a detergent bound open conformation | | 分子名称: | 1,2-ETHANEDIOL, 3,6,12,15,18,21,24-HEPTAOXAHEXATRIACONTAN-1-OL, FE (III) ION, ... | | 著者 | Wild, K, Sinning, I. | | 登録日 | 2023-08-30 | | 公開日 | 2024-10-30 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Vitis vinifera Lipoxygenase LoxA is an Allosteric Dimer Activated by Lipidic Surfaces.
J.Mol.Biol., 436, 2024
|
|
8VK4
 
 | |
5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | 分子名称: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | 著者 | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | 登録日 | 2016-09-15 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
6DLS
 
 | |
9BHO
 
 | | Crystal structure of KRAS G12S in a transition state mimetic complex with CYPA and RMC-7977 | | 分子名称: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | 登録日 | 2024-04-21 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Pharmacological restoration of GTP hydrolysis by mutant RAS.
Nature, 637, 2025
|
|
9BGH
 
 | | Crystal structure of KRAS G12D in a transition state mimetic complex with CYPA and RMC-7977 | | 分子名称: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, ALUMINUM FLUORIDE, GTPase KRas, ... | | 著者 | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | 登録日 | 2024-04-18 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Pharmacological restoration of GTP hydrolysis by mutant RAS.
Nature, 637, 2025
|
|
5KWB
 
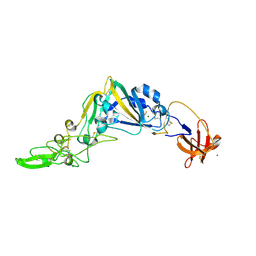 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | 著者 | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2016-07-17 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
9BI2
 
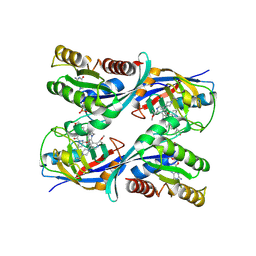 | | Crystal structure of GMPPNP bound KRAS G12C in complex with CYPA and RMC-7977 | | 分子名称: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | 著者 | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | 登録日 | 2024-04-22 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Pharmacological restoration of GTP hydrolysis by mutant RAS.
Nature, 637, 2025
|
|
5CEX
 
 | |
9BHQ
 
 | | Crystal structure of KRAS G12A in a transition state mimetic complex with CYPA and RMC-7977 | | 分子名称: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | 登録日 | 2024-04-21 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pharmacological restoration of GTP hydrolysis by mutant RAS.
Nature, 637, 2025
|
|
9BHP
 
 | | Crystal structure of KRAS G12C in a transition state mimetic complex with CYPA and RMC-7977 | | 分子名称: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Pourfarjam, Y, Goldgur, Y, Cuevas-Navarro, A, Lito, P. | | 登録日 | 2024-04-21 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pharmacological restoration of GTP hydrolysis by mutant RAS.
Nature, 637, 2025
|
|
8PY8
 
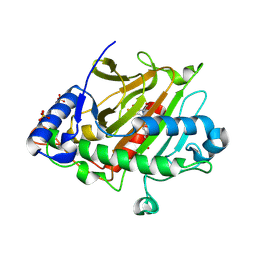 | |
5B3G
 
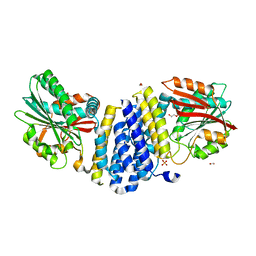 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Hirano, Y, Nakagawa, M, Hakoshima, T. | | 登録日 | 2016-02-29 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5CP8
 
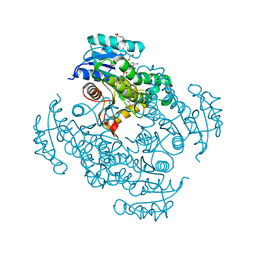 | | The effect of isoleucine to alanine mutation on InhA enzyme crystallization pattern and substrate binding loop conformation and flexibility | | 分子名称: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, ... | | 著者 | Li, H.-J, Lai, C.-T, Liu, N, Yu, W, Shah, S, Bommineni, G.R, Perrone, V, Garcia-Diaz, M, Tonge, P.J, Simmerling, C. | | 登録日 | 2015-07-21 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rational Modulation of the Induced-Fit Conformational Change for Slow-Onset Inhibition in Mycobacterium tuberculosis InhA.
Biochemistry, 54, 2015
|
|
8PY9
 
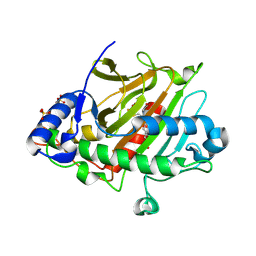 | | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure | | 分子名称: | (2S)-2-azanyl-6-[[(2R)-1-[[(2R)-3-methyl-1-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-3-sulfanylidene-propan-2-yl]amino]-6-oxidanylidene-hexanoic acid, FE (III) ION, Isopenicillin N synthase, ... | | 著者 | Rabe, P, Schofield, C.J. | | 登録日 | 2023-07-25 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Isopenicillin N synthase in complex with Fe, O2, ACdV or thioaldehyde after 3s O2 exposure
To Be Published
|
|
6G4P
 
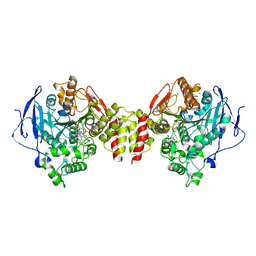 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(~{Z})-hydroxyiminomethyl]pyridin-3-ol, Acetylcholinesterase, ... | | 著者 | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | 登録日 | 2018-03-28 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
5IXS
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 9: (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one | | 分子名称: | (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Ultsch, M, Eigenbrot, C. | | 登録日 | 2016-03-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
6DKE
 
 | |
9GV2
 
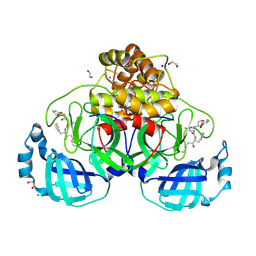 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalently bound inhibitor FP237 (compound 8p in publication) | | 分子名称: | (2S)-2-[2-(3-methoxyphenoxy)ethanoylamino]-4-methyl-N-[(2S)-3-oxidanylidene-1-phenyl-pentan-2-yl]pentanamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Strater, N, Sylvester, K, Muller, C.E, Flury, P, Kruger, N, Breidenbach, J, Guetschow, M, Laufer, S.A, Pillaiyar, T. | | 登録日 | 2024-09-20 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Design, Synthesis, and Unprecedented Interactions of Covalent Dipeptide-Based Inhibitors of SARS-CoV-2 Main Protease and Its Variants Displaying Potent Antiviral Activity.
J.Med.Chem., 68, 2025
|
|
3I52
 
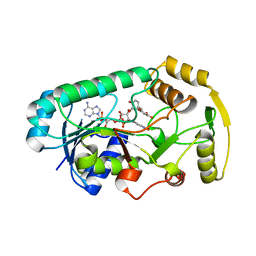 | | Ternary complex structure of leucoanthocyanidin reductase from vitis vinifera | | 分子名称: | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative leucoanthocyanidin reductase 1 | | 著者 | Mauge, C, Gargouri, M, d'Estaintot, B.L, Granier, T, Gallois, B. | | 登録日 | 2009-07-03 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera.
J.Mol.Biol., 397, 2010
|
|
9FTR
 
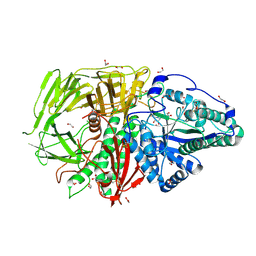 | | Drosophila golgi alpha-mannosidase II (dGMII) in complex with amide modified swainsonine-configured alkyl indolizidine | | 分子名称: | 1,2-ETHANEDIOL, Alpha-mannosidase 2, ZINC ION, ... | | 著者 | Bennett, M, Koemans, T, Overkleeft, H.S, Davies, G.J. | | 登録日 | 2024-06-25 | | 公開日 | 2024-10-09 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure-guided design of C3-branched swainsonine as potent and selective human Golgi alpha-mannosidase (GMII) inhibitor.
Chem.Commun.(Camb.), 60, 2024
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | 著者 | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2018-09-25 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | 著者 | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | 登録日 | 2018-09-25 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6FQT
 
 | |
8HPE
 
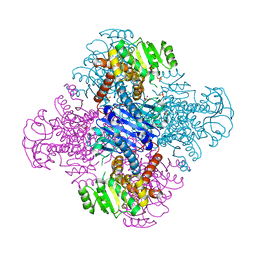 | | Crystal structure of Leucine dehydrogenase | | 分子名称: | GLYCEROL, Leucine dehydrogenase, SULFATE ION | | 著者 | Li, X, Song, W. | | 登録日 | 2022-12-12 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | A Tri-Enzyme Cascade for Efficient Production of L-2-Aminobutyrate from L-Threonine.
Chembiochem, 24, 2023
|
|
