7O31
 
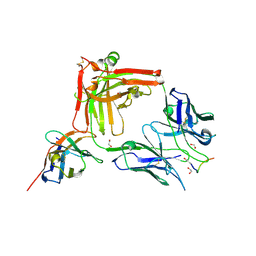 | | Crystal structure of the anti-PAS Fab 1.2 in complex with its epitope peptide and the anti-Kappa VHH domain | | 分子名称: | 1,2-ETHANEDIOL, PAS#1 epitope peptide, anti-Kappa VHH domain, ... | | 著者 | Schilz, J, Schiefner, A, Skerra, A. | | 登録日 | 2021-04-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular recognition of structurally disordered Pro/Ala-rich sequences (PAS) by antibodies involves an Ala residue at the hot spot of the epitope.
J.Mol.Biol., 433, 2021
|
|
8UK9
 
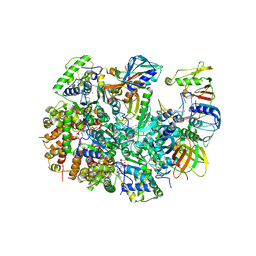 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | 著者 | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | 登録日 | 2023-10-12 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6DP2
 
 | |
7NZ4
 
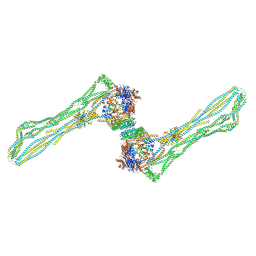 | | Cryo-EM structure of the MukBEF dimer | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, Chromosome partition protein MukB, ... | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2021-03-23 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (13 Å) | | 主引用文献 | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
8UNH
 
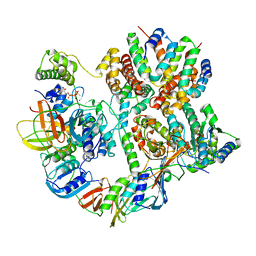 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | 著者 | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | 登録日 | 2023-10-19 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6DPO
 
 | |
8U0V
 
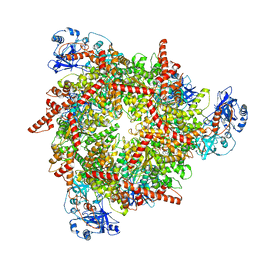 | | S. cerevisiae Pex1/Pex6 with 1 mM ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Peroxisomal ATPase PEX1, Peroxisomal ATPase PEX6 | | 著者 | Gardner, B.M. | | 登録日 | 2023-08-29 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | ELECTRON MICROSCOPY (3.89 Å) | | 主引用文献 | The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
J.Biol.Chem., 300, 2023
|
|
6DQ4
 
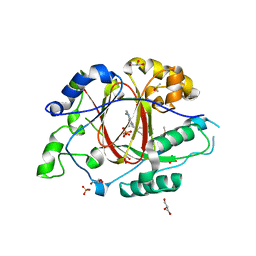 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | 分子名称: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-06-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.392 Å) | | 主引用文献 | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
7OGL
 
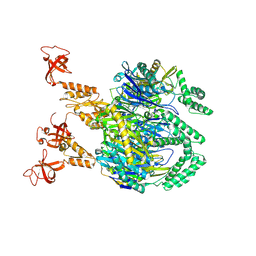 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase | | 分子名称: | Polyribonucleotide nucleotidyltransferase | | 著者 | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | 登録日 | 2021-05-06 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
8UC1
 
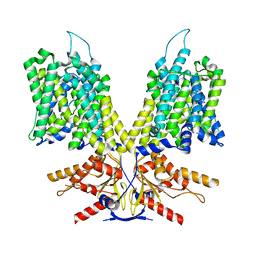 | |
8GK8
 
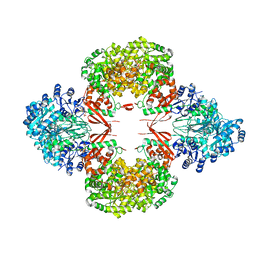 | | R21A Staphylococcus aureus pyruvate carboxylase | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, COENZYME A, ... | | 著者 | Laseke, A.J, St.Maurice, M. | | 登録日 | 2023-03-17 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Allosteric Site at the Biotin Carboxylase Dimer Interface Mediates Activation and Inhibition in Staphylococcus aureus Pyruvate Carboxylase.
Biochemistry, 62, 2023
|
|
6DQV
 
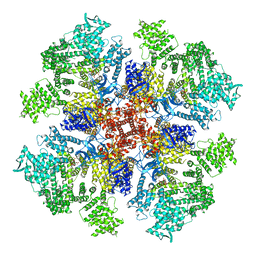 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | 著者 | Hite, R.K, Paknejad, N. | | 登録日 | 2018-06-11 | | 公開日 | 2018-08-01 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7O2C
 
 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH the benzodiazepine-based probe BAY-6035 | | 分子名称: | (2S)-1-[[(1R,5S)-3-azabicyclo[3.1.0]hexan-3-yl]carbonyl]-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | 著者 | Steuber, H. | | 登録日 | 2021-03-30 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
7OGM
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)-Hfq | | 分子名称: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase, RNA-binding protein Hfq | | 著者 | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | 登録日 | 2021-05-06 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7NYW
 
 | |
7NZ2
 
 | | Cryo-EM structure of the MukBEF-MatP-DNA tetrad | | 分子名称: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | 著者 | Buermann, F, Lowe, J. | | 登録日 | 2021-03-23 | | 公開日 | 2021-07-07 | | 最終更新日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites.
Mol.Cell, 81, 2021
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | 著者 | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6DKT
 
 | |
8V4G
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | 著者 | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | 登録日 | 2023-11-29 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
7OGK
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ) | | 分子名称: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase | | 著者 | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | 登録日 | 2021-05-06 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7NT6
 
 | | CryoEM structure of the Nipah virus nucleocapsid spiral clam-shaped assembly | | 分子名称: | Nucleoprotein, RNA (42-MER), RNA (48-MER) | | 著者 | Ker, D.S, Jenkins, H.T, Greive, S.J, Antson, A.A. | | 登録日 | 2021-03-09 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | CryoEM structure of the Nipah virus nucleocapsid assembly.
Plos Pathog., 17, 2021
|
|
4N0G
 
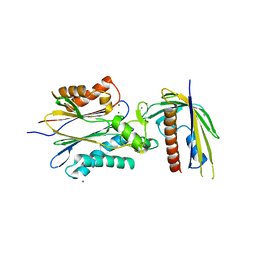 | | Crystal Structure of PYL13-PP2CA complex | | 分子名称: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | 著者 | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | 登録日 | 2013-10-01 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
7NYZ
 
 | |
4MZN
 
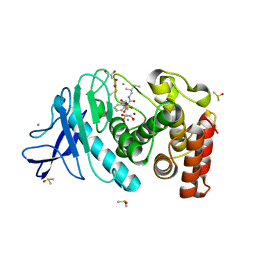 | | Thermolysin in complex with UBTLN59 | | 分子名称: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Krimmer, S.G, Heine, A, Klebe, G. | | 登録日 | 2013-09-30 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.172 Å) | | 主引用文献 | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 4, 2014
|
|
6DLV
 
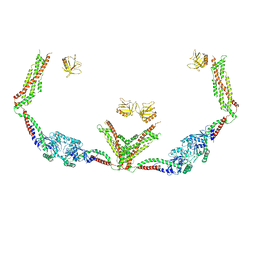 | | Cryo-EM of the GTP-bound human dynamin-1 polymer assembled on the membrane in the super constricted state | | 分子名称: | Dynamin-1 | | 著者 | Kong, L, Wang, H, Fang, S, Canagarajah, B, Kehr, A.D, Rice, W.J, Hinshaw, J.E. | | 登録日 | 2018-06-02 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (10.1 Å) | | 主引用文献 | Cryo-EM of the dynamin polymer assembled on lipid membrane.
Nature, 560, 2018
|
|
