4R3O
 
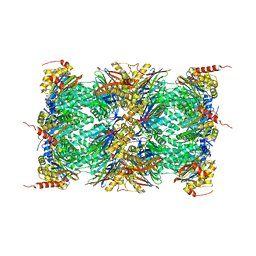 | | Human Constitutive 20S Proteasome | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Sacchettini, J.C, Harshbarger, W.H. | | 登録日 | 2014-08-16 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
4MDS
 
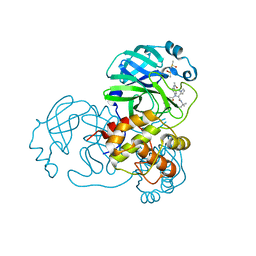 | | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and non-covalent nanomolar inhibitors with an induced-fit binding | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[4-(acetylamino)phenyl]-2-(1H-benzotriazol-1-yl)-N-[(1R)-2-[(2-methylbutan-2-yl)amino]-1-(1-methyl-1H-pyrrol-2-yl)-2-oxoethyl]acetamide | | 著者 | Mesecar, A.D, Grum-Tokars, V. | | 登録日 | 2013-08-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5CDZ
 
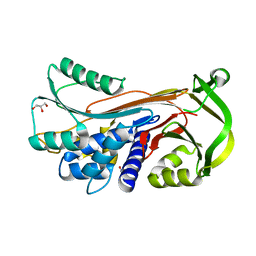 | | Crystal structure of conserpin in the latent state | | 分子名称: | Conserpin in the latent state, GLYCEROL | | 著者 | Porebski, B.T, McGowan, S, Keleher, S, Buckle, A.M. | | 登録日 | 2015-07-06 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.449 Å) | | 主引用文献 | Smoothing a rugged protein folding landscape by sequence-based redesign.
Sci Rep, 6, 2016
|
|
5CE0
 
 | |
5CDX
 
 | |
6UTU
 
 | | Crystal structure of minor pseudopilin ternary complex of XcpVWX from the Type 2 secretion system of Pseudomonas aeruginosa in the P3 space group | | 分子名称: | CALCIUM ION, Type II secretion system protein I, Type II secretion system protein J, ... | | 著者 | Zhang, Y, Wang, S, Jia, Z. | | 登録日 | 2019-10-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | In Situ Proteolysis Condition-Induced Crystallization of the XcpVWX Complex in Different Lattices.
Int J Mol Sci, 21, 2020
|
|
4U13
 
 | | Crystal structure of putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution | | 分子名称: | putative polyketide cyclase SMa1630 | | 著者 | Shabalin, I.G, Bacal, P, Osinski, T, Cooper, D.R, Szlachta, K, Stead, M, Grabowski, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-07-14 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a putative polyketide cyclase (protein SMa1630) from Sinorhizobium meliloti at 2.3 A resolution
to be published
|
|
4WJI
 
 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-09-30 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
4XK2
 
 | | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666 | | 分子名称: | Aldo/keto reductase, CHLORIDE ION, SODIUM ION | | 著者 | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Sroka, P, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2015-01-09 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666
to be published
|
|
2HGO
 
 | | NMR structure of Cassiicolin | | 分子名称: | 1,5-anhydro-3-O-methyl-D-mannitol, CASSIICOLIN | | 著者 | Barthe, P, Pujade-Renault, V, Roumestand, C, de Lamotte, F. | | 登録日 | 2006-06-27 | | 公開日 | 2007-02-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of Cassiicolin, a Host-selective Protein Toxin from Corynespora cassiicola
J.Mol.Biol., 367, 2007
|
|
7TOB
 
 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
1HTM
 
 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | 分子名称: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | 著者 | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | 登録日 | 1994-11-02 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
1I1B
 
 | |
1GUR
 
 | | GURMARIN, A SWEET TASTE-SUPPRESSING POLYPEPTIDE, NMR, 10 STRUCTURES | | 分子名称: | GURMARIN | | 著者 | Arai, K, Ishima, R, Morikawa, S, Imoto, T, Yoshimura, S, Aimoto, S, Akasaka, K. | | 登録日 | 1996-03-12 | | 公開日 | 1996-08-01 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of gurmarin, a sweet taste-suppressing polypeptide.
J.Biomol.NMR, 5, 1995
|
|
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | 分子名称: | GAMMA-1-P THIONIN | | 著者 | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | 登録日 | 1992-07-29 | | 公開日 | 1993-10-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | 分子名称: | H-NS | | 著者 | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | 登録日 | 1995-04-06 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HNS
 
 | | H-NS (DNA-BINDING DOMAIN) | | 分子名称: | H-NS | | 著者 | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | 登録日 | 1995-04-06 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1IRH
 
 | | The Solution Structure of The Third Kunitz Domain of Tissue Factor Pathway Inhibitor | | 分子名称: | tissue factor pathway inhibitor | | 著者 | Mine, S, Yamazaki, T, Miyata, T, Hara, S, Kato, H. | | 登録日 | 2001-10-02 | | 公開日 | 2002-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor.
Biochemistry, 41, 2002
|
|
1KTH
 
 | |
5DHM
 
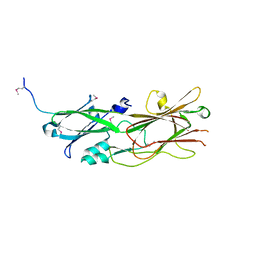 | |
5EMN
 
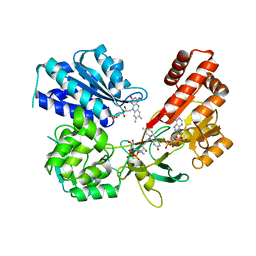 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase(A287P mutant) | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | 登録日 | 2015-11-06 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5FA6
 
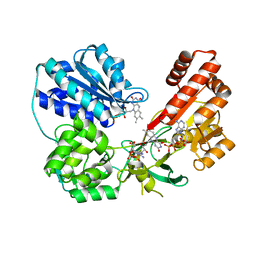 | | wild type human CYPOR | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | 登録日 | 2015-12-11 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
2D98
 
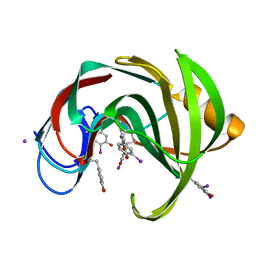 | | Structure of VIL (extra KI/I2 added)-xylanase | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Miyatake, H, Hasegawa, T, Yamano, A. | | 登録日 | 2005-12-09 | | 公開日 | 2006-07-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1PCA
 
 | | THREE DIMENSIONAL STRUCTURE OF PORCINE PANCREATIC PROCARBOXYPEPTIDASE A. A COMPARISON OF THE A AND B ZYMOGENS AND THEIR DETERMINANTS FOR INHIBITION AND ACTIVATION | | 分子名称: | CITRIC ACID, PROCARBOXYPEPTIDASE A PCPA, VALINE, ... | | 著者 | Guasch, A, Coll, M, Aviles, F.X, Huber, R. | | 登録日 | 1991-10-28 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation.
J.Mol.Biol., 224, 1992
|
|
1QY6
 
 | | Structue of V8 Protease from Staphylococcus aureus | | 分子名称: | POTASSIUM ION, serine protease | | 著者 | Prasad, L, Leduc, Y, Hayakawa, K, Delbaere, L.T.J. | | 登録日 | 2003-09-09 | | 公開日 | 2004-02-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
