2EX4
 
 | | Crystal Structure of Human methyltransferase AD-003 in complex with S-adenosyl-L-homocysteine | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, adrenal gland protein AD-003 | | 著者 | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | 登録日 | 2005-11-07 | | 公開日 | 2005-11-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Crystal Structure of Human AD-003 protein in complex with S-adenosyl-L-homocysteine
To be Published
|
|
2EX5
 
 | |
2EX6
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX8
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-G | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX9
 
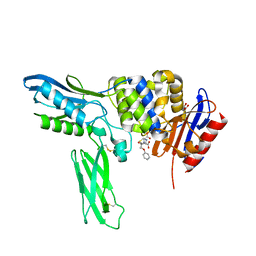 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-V | | 分子名称: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXA
 
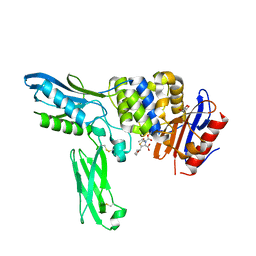 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FAROM | | 分子名称: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXB
 
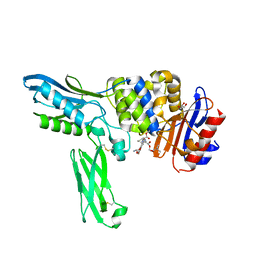 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FLOMOX | | 分子名称: | 2,2-dimethylpropanoyloxymethyl (2R)-5-(aminocarbonyloxymethyl)-2-[(1R)-1-[[(Z)-2-(2-azanyl-1,3-thiazol-4-yl)pent-2-enoyl]amino]-2-oxidanylidene-ethyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylate, GLYCEROL, Penicillin-binding protein 4 | | 著者 | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-06-13 | | 最終更新日 | 2016-10-19 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXC
 
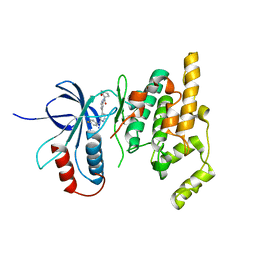 | | Inhibitor complex of JNK3 | | 分子名称: | Mitogen-activated protein kinase 10, N-{2'-[(4-FLUOROPHENYL)AMINO]-4,4'-BIPYRIDIN-2-YL}-4-METHOXYCYCLOHEXANECARBOXAMIDE | | 著者 | Xue, Y. | | 登録日 | 2005-11-08 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Inhibitor complex of JNK3
To be Published
|
|
2EXD
 
 | | The solution structure of the C-terminal domain of a nfeD homolog from Pyrococcus horikoshii | | 分子名称: | nfeD short homolog | | 著者 | Kuwahara, Y, Ohno, A, Morii, T, Tochio, H, Shirakawa, M, Hiroaki, H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the C-terminal domain of NfeD reveals a novel membrane-anchored OB-fold.
Protein Sci., 17, 2008
|
|
2EXE
 
 | | Crystal structure of the phosphorylated CLK3 | | 分子名称: | Dual specificity protein kinase CLK3 | | 著者 | Papagrigoriou, E, Rellos, P, Das, S, Bullock, A, Ball, L.J, Turnbull, A, Savitsky, P, Fedorov, O, Johansson, C, Ugochukwu, E, Sobott, F, von Delft, F, Edwards, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2005-11-08 | | 公開日 | 2005-11-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the phosphorylated CLK3
TO BE PUBLISHED
|
|
2EXF
 
 | |
2EXG
 
 | | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain | | 分子名称: | (5R)-2-SULFANYL-5-[4-(TRIFLUOROMETHYL)BENZYL]-1,3-THIAZOL-4-ONE, Afadin | | 著者 | Joshi, M, Vargas, C, Boisguerin, P, Krause, G, Schade, M, Oschkinat, H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-10-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
2EXH
 
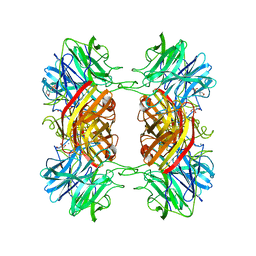 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXI
 
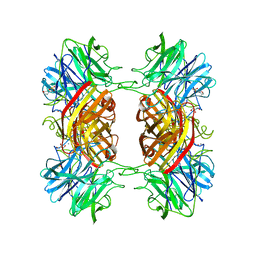 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXJ
 
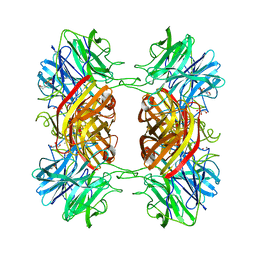 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXK
 
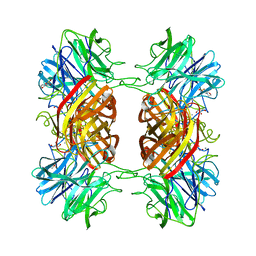 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXL
 
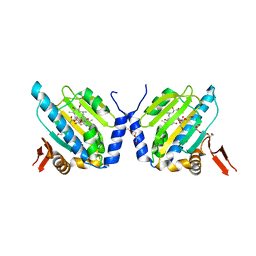 | | GRP94 N-terminal Domain bound to geldanamycin | | 分子名称: | Endoplasmin, GELDANAMYCIN, PENTAETHYLENE GLYCOL, ... | | 著者 | Reardon, P.N, Immormino, R.M, Gewirth, D.T. | | 登録日 | 2005-11-08 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
2EXM
 
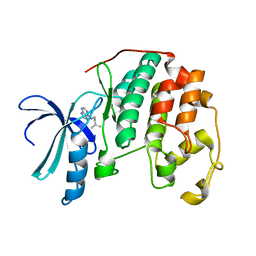 | | Human CDK2 in complex with isopentenyladenine | | 分子名称: | Cell division protein kinase 2, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE | | 著者 | Schulze-Gahmen, U. | | 登録日 | 2005-11-08 | | 公開日 | 2005-12-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Multiple modes of ligand recognition: crystal structures of cyclin-dependent protein kinase 2 in complex with ATP and two inhibitors, olomoucine and isopentenyladenine.
Proteins, 22, 1995
|
|
2EXN
 
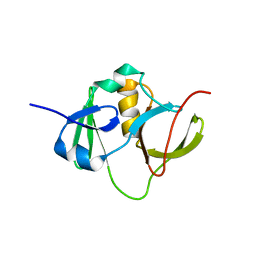 | | Solution structure for the protein coded by gene locus BB0938 of Bordetella bronchiseptica. Northeast Structural Genomics target BoR11. | | 分子名称: | Hypothetical protein BoR11 | | 著者 | Rossi, P, Ramelot, T, Xiao, R, Ho, C.K, Ma, L.-C, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2005-11-08 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | (1)H, (13)C, and (15)N Resonance Assignments for the Protein Coded by Gene Locus BB0938 of Bordetella bronchiseptica
J.Biomol.NMR, 33, 2005
|
|
2EXO
 
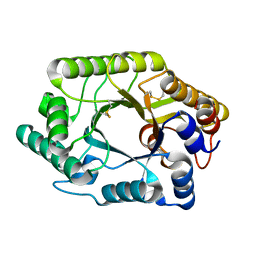 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE BETA-1,4-GLYCANASE CEX FROM CELLULOMONAS FIMI | | 分子名称: | EXO-1,4-BETA-D-GLYCANASE | | 著者 | White, A, Withers, S.G, Gilkes, N.R, Rose, D.R. | | 登録日 | 1994-07-11 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the catalytic domain of the beta-1,4-glycanase cex from Cellulomonas fimi.
Biochemistry, 33, 1994
|
|
2EXR
 
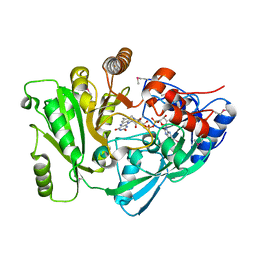 | | X-Ray Structure of Cytokinin Oxidase/Dehydrogenase (CKX) From Arabidopsis Thaliana AT5G21482 | | 分子名称: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2005-11-08 | | 公開日 | 2005-11-29 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
2EXS
 
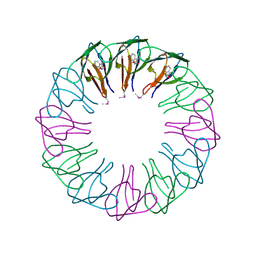 | | TRAP3 (engineered TRAP) | | 分子名称: | TRYPTOPHAN, Transcription attenuation protein mtrB | | 著者 | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-08-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
2EXT
 
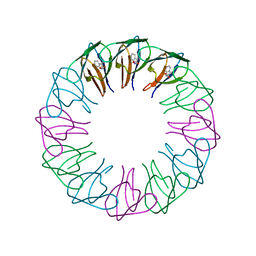 | | TRAP4 (engineered TRAP) | | 分子名称: | TRYPTOPHAN, Transcription attenuation protein mtrB | | 著者 | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | 登録日 | 2005-11-08 | | 公開日 | 2006-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
2EXU
 
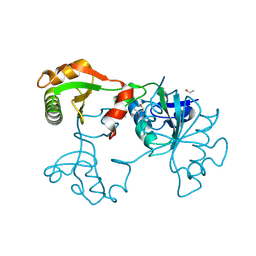 | | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHANOL, Transcription initiation protein SPT4/SPT5, ... | | 著者 | Xu, F, Guo, M, Fang, P, Teng, M, Niu, L. | | 登録日 | 2005-11-08 | | 公開日 | 2006-11-08 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal Structure of Saccharomyces cerevisiae transcription elongation factors Spt4-Spt5NGN domain
To be published
|
|
2EXV
 
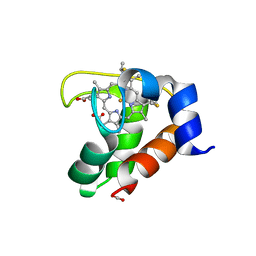 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | 分子名称: | ACETIC ACID, Cytochrome c-551, HEME C | | 著者 | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | 登録日 | 2005-11-09 | | 公開日 | 2006-02-07 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
