7YUE
 
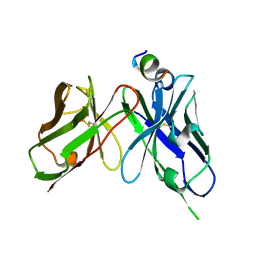 | | Epitope-directed anti-SARS CoV 2 scFv engineered against the key spike protein region. | | 分子名称: | Single chain variable Fragment, Spike protein S2 | | 著者 | Kumar, U, Jaiswal, D, Gaur, V, Salunke, D.M. | | 登録日 | 2022-08-17 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Epitope-directed anti-SARS-CoV-2 scFv engineered against the key spike protein region could block membrane fusion.
Protein Sci., 32, 2023
|
|
8E26
 
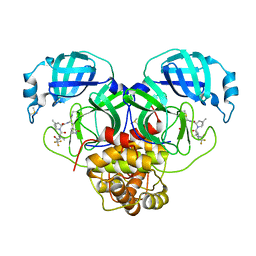 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Godoy, A.S, Oliva, G. | | 登録日 | 2022-08-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E25
 
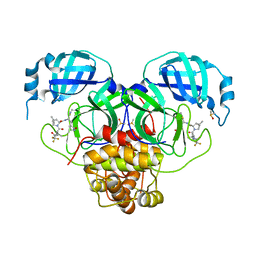 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | 著者 | Noske, G.D, Godoy, A.S, Oliva, G. | | 登録日 | 2022-08-14 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7YSX
 
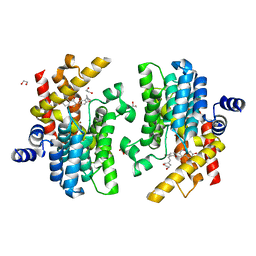 | | Crystal structure of PDE4D complexed with licoisoflavone A | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2022-08-13 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E1Y
 
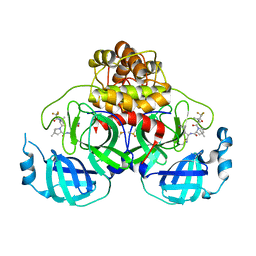 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7YQF
 
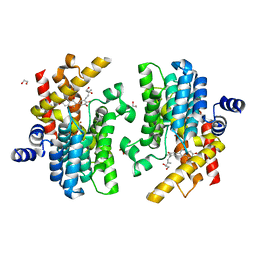 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2022-08-06 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DZ9
 
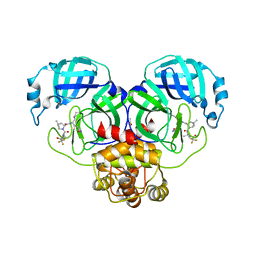 | | Crystal Structure of SARS-CoV-2 Main protease G143S mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.664 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ0
 
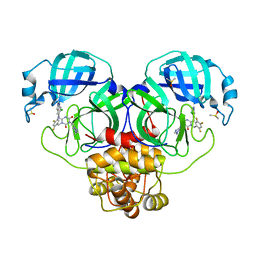 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZA
 
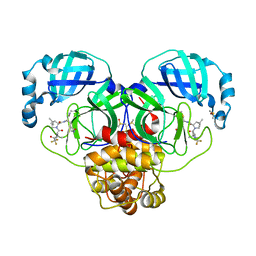 | | Crystal Structure of SARS-CoV-2 Main protease A193T mutant in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.961 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ6
 
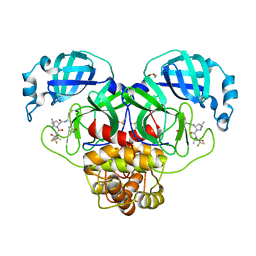 | | Crystal Structure of SARS-CoV-2 Main protease mutant Q189K in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.366 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ1
 
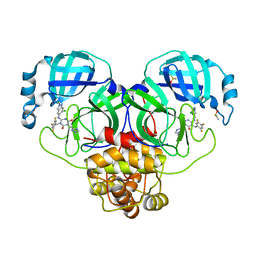 | | Crystal Structure of SARS-CoV-2 Main protease mutant M49I in complex with Ensitrelvir | | 分子名称: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ2
 
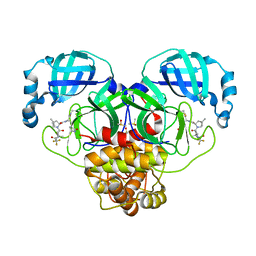 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.129 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZB
 
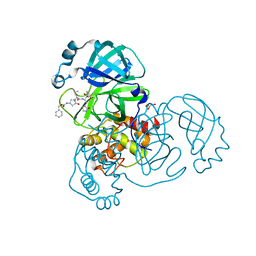 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 11 | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, benzyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-5-oxopyrrolidin-3-yl}carbamate | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DZC
 
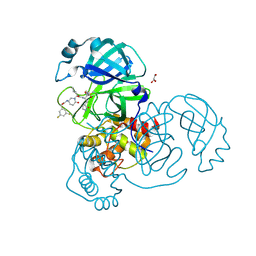 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor 17 | | 分子名称: | (3,5-difluorophenyl)methyl {(3S)-1-[(2S)-1-({(2S,3R)-4-(cyclopropylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-6-oxopiperidin-3-yl}carbamate, 3C-like proteinase nsp5, GLYCEROL | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2022-08-06 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained alpha , gamma-AA Peptide Scaffold.
Chemistry, 29, 2023
|
|
8DYA
 
 | |
8DXS
 
 | |
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | 分子名称: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | 分子名称: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | 分子名称: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
8DW9
 
 | |
8DWA
 
 | |
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | 分子名称: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | 著者 | Kwon, Y.D, Gorman, J, Kwong, P.D. | | 登録日 | 2022-07-30 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (4.11 Å) | | 主引用文献 | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
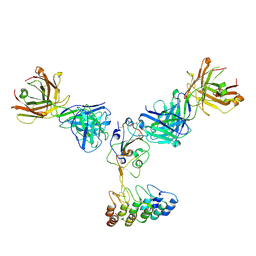 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | 分子名称: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | 著者 | Kwon, Y.D, Gorman, J, Kwong, P.D. | | 登録日 | 2022-07-30 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (4.26 Å) | | 主引用文献 | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8AJL
 
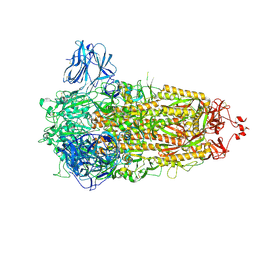 | | Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | 登録日 | 2022-07-28 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
8AJA
 
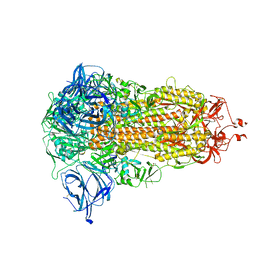 | | Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Hueting, D, Schriever, K, Wallden, K, Andrell, J, Syren, P.O. | | 登録日 | 2022-07-27 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Design, structure and plasma binding of ancestral beta-CoV scaffold antigens.
Nat Commun, 14, 2023
|
|
