8JDM
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (rotated state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDL
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.42 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDK
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | 登録日 | 2023-05-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDI
 
 | |
8JCZ
 
 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCY
 
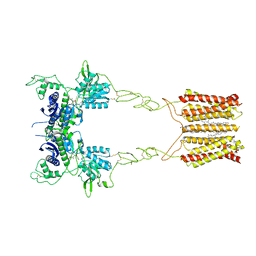 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
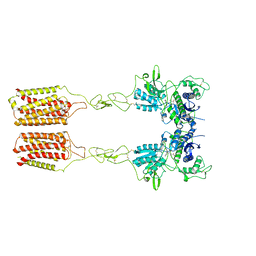 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCW
 
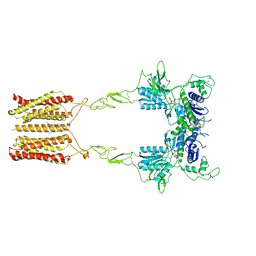 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode I) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCV
 
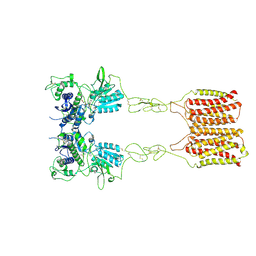 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCU
 
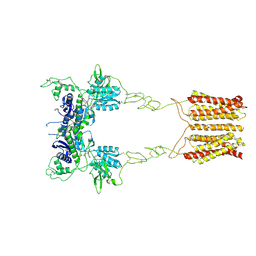 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | 分子名称: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | 著者 | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | 登録日 | 2023-05-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCR
 
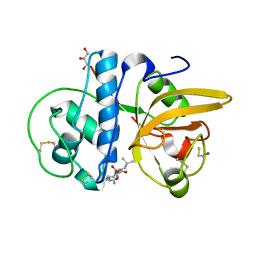 | | Crystal structure of Procerain from Calotropis gigantea (pH 6.0) | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | 著者 | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of Procerain from Calotropis gigantea
To Be Published
|
|
8JCN
 
 | |
8JCM
 
 | |
8JCK
 
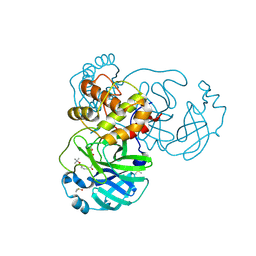 | |
8JCJ
 
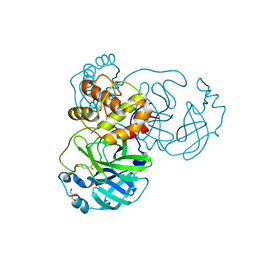 | |
8JCA
 
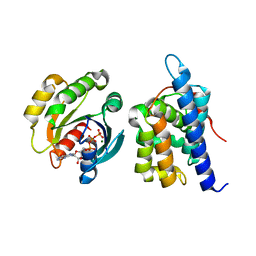 | |
8JC5
 
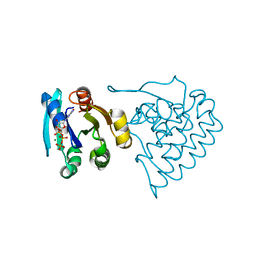 | |
8JBR
 
 | | Structure of McyA2-CAPCP | | 分子名称: | 4'-PHOSPHOPANTETHEINE, ADENOSINE-5'-TRIPHOSPHATE, McyA protein | | 著者 | Peng, Y.J. | | 登録日 | 2023-05-09 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Modular catalytic activity of nonribosomal peptide synthetases depends on the dynamic interaction between adenylation and condensation domains.
Structure, 32, 2024
|
|
8JBP
 
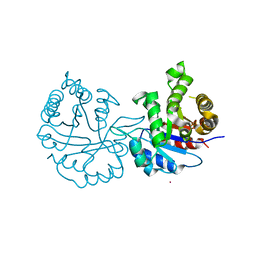 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.45 angstroms resolution with an arsenic atom bound at Cys57 | | 分子名称: | ARSENIC, Triosephosphate isomerase | | 著者 | Kuaprasert, B, Leartsakulpanich, U, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Robinson, R.C, Zhou, Y, Mungthin, M, Leelayoova, S, Saehlee, S, Choowongkomon, K. | | 登録日 | 2023-05-09 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
8JBO
 
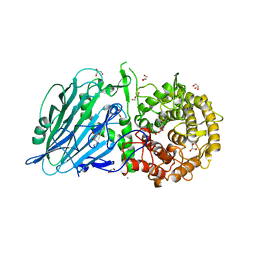 | | Crystal structure of TxGH116 from Thermoanaerobacterium xylanolyticum with isofagomine | | 分子名称: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | 著者 | Pengthaisong, S, Ketudat Cairns, J.R. | | 登録日 | 2023-05-09 | | 公開日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for inhibition of a GH116 beta-glucosidase and its missense mutants by GBA2 inhibitors: Crystallographic and quantum chemical study.
Chem.Biol.Interact., 384, 2023
|
|
8JBM
 
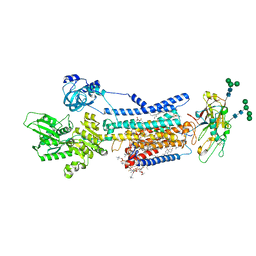 | | Crystal structure of Na+,K+-ATPase in the E1.Mn2+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBL
 
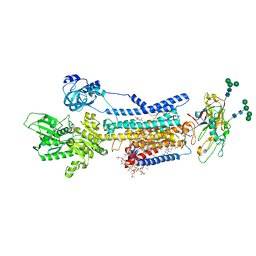 | | Crystal structure of Na+,K+-ATPase in the E1.Mg2+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBK
 
 | | Crystal structure of Na+,K+-ATPase in the E1.3Na+ state | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kanai, R, Vilsen, B, Cornelius, F, Toyoshima, C. | | 登録日 | 2023-05-09 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of Na + ,K + -ATPase reveal the mechanism that converts the K + -bound form to Na + -bound form and opens and closes the cytoplasmic gate.
Febs Lett., 597, 2023
|
|
8JBA
 
 | |
