8WHA
 
 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
 | | Structure of DDM1-nucleosome complex in the apo state | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
 | | Structure of nucleosome core particle of Arabidopsis thaliana | | 分子名称: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-23 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
6TXC
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dCMPNPP and Mg | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | 著者 | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A, Kelly, G, Taylor, I.A. | | 登録日 | 2020-01-14 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TXF
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mn | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | 著者 | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | 登録日 | 2020-01-14 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6DT3
 
 | | 1.2 Angstrom Resolution Crystal Structure of Nucleoside Triphosphatase NudI from Klebsiella pneumoniae in Complex with HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside triphosphatase NudI | | 著者 | Minasov, G, Shuvalova, L, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-15 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7K6S
 
 | | Crystal structure of the bromodomain (BD) of human Bromodomain and PHD finger-containing Transcription Factor (BPTF) bound to Compound 4 (SKT1174) | | 分子名称: | (7-amino-3,4-dihydroquinolin-1(2H)-yl)(cyclopropyl)methanone, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Chan, A, Schonbrunn, E. | | 登録日 | 2020-09-21 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | New inhibitors for the BPTF bromodomain enabled by structural biology and biophysical assay development.
Org.Biomol.Chem., 18, 2020
|
|
8T5L
 
 | |
7KDW
 
 | |
3C6T
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 14 | | 分子名称: | 2-[3-chloro-5-(3-chloro-5-cyanophenoxy)phenoxy]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase | | 著者 | Yan, Y, Prasad, S. | | 登録日 | 2008-02-05 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8U6L
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((6-cyanonaphthalen-1-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ748), a non-nucleoside inhibitor | | 分子名称: | MAGNESIUM ION, N-(2-{5-chloro-2-[(6-cyanonaphthalen-1-yl)oxy]phenoxy}ethyl)-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, ... | | 著者 | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2023-09-13 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.486 Å) | | 主引用文献 | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
3C6U
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 22 | | 分子名称: | 3-chloro-5-[2-chloro-5-(1H-indazol-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase | | 著者 | Yan, Y, Prasad, S. | | 登録日 | 2008-02-05 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6EOR
 
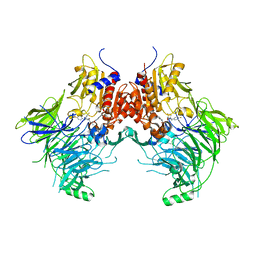 | | DPP9 - 1G244 | | 分子名称: | (2~{S})-2-azanyl-4-[4-[bis(4-fluorophenyl)methyl]piperazin-1-yl]-1-(1,3-dihydroisoindol-2-yl)butane-1,4-dione, Dipeptidyl peptidase 9 | | 著者 | Ross, B.R, Huber, R. | | 登録日 | 2017-10-10 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EOQ
 
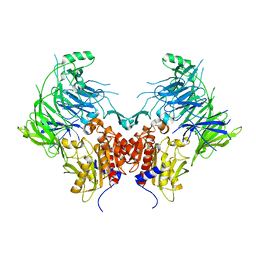 | | DPP9 - Apo | | 分子名称: | Dipeptidyl peptidase 9 | | 著者 | Ross, B.R, Huber, R. | | 登録日 | 2017-10-10 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FFZ
 
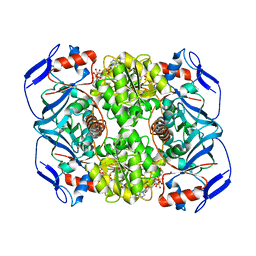 | | Crystal structure of R. ruber ADH-A, mutant F43H, Y54L | | 分子名称: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | 登録日 | 2018-01-09 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
8X2Z
 
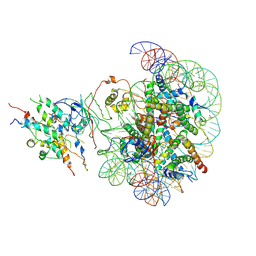 | | The class2 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | 分子名称: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | 著者 | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | 登録日 | 2023-11-10 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2Y
 
 | | The class1 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | 分子名称: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | 著者 | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | 登録日 | 2023-11-10 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2X
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex at pre-H4-acetylation state | | 分子名称: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | 著者 | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | 登録日 | 2023-11-10 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X31
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex with Ac-CoA at resetting state | | 分子名称: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | 著者 | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | 登録日 | 2023-11-10 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-04-02 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9CPC
 
 | | Atomic model of porcine brain ventricles cilia doublet microtubule (48-nm periodicity) | | 分子名称: | Armadillo repeat-containing protein 4 isoform 1, CFAP107/C1orf158, Chromosome 1 C15orf65 homolog, ... | | 著者 | Sun, C, Zeng, J, Zhang, R. | | 登録日 | 2024-07-18 | | 公開日 | 2024-10-23 | | 最終更新日 | 2025-03-19 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural diversity of axonemes across mammalian motile cilia.
Nature, 637, 2025
|
|
8CWW
 
 | | Structure of S. cerevisiae Hop1 CBR bound to a nucleosome | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | 登録日 | 2022-05-19 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
8CZE
 
 | | Structure of a Xenopus Nucleosome with Widom 601 DNA | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | 登録日 | 2022-05-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
9EH1
 
 | |
6W5I
 
 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | 登録日 | 2020-03-13 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
