2N3W
 
 | |
2Y00
 
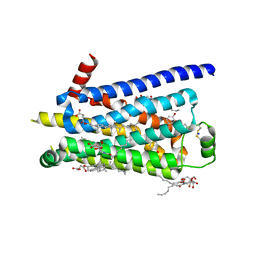 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST DOBUTAMINE (CRYSTAL DOB92) | | 分子名称: | BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, DOBUTAMINE, ... | | 著者 | Warne, A, Moukhametzianov, R, Baker, J.G, Nehme, R, Edwards, P.C, Leslie, A.G.W, Schertler, G.F.X, Tate, C.G. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor
Nature, 469, 2011
|
|
7QYR
 
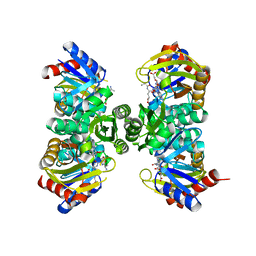 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | 著者 | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | 登録日 | 2022-01-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
8PJ8
 
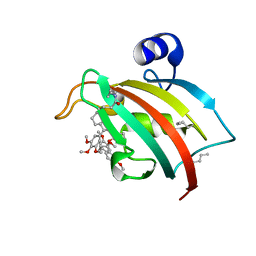 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | 分子名称: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Meyners, C, Charalampidou, A, Hausch, F. | | 登録日 | 2023-06-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
7JQ0
 
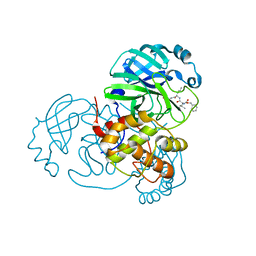 | |
2N4F
 
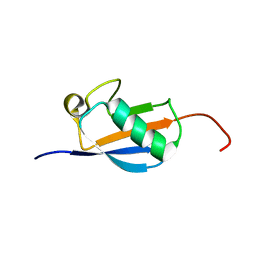 | | EC-NMR Structure of Arabidopsis thaliana At2g32350 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AR3433A | | 分子名称: | uncharacterized protein AR3433A | | 著者 | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-06-17 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
8PYR
 
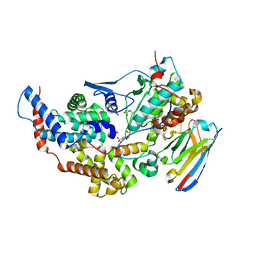 | | Crystal structure of the dual T-loop phosphorylated Cdk7/CycH/Mat1 complex | | 分子名称: | 1,2-ETHANEDIOL, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | 著者 | Anand, K, Duster, R, Geyer, M. | | 登録日 | 2023-07-26 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis of Cdk7 activation by dual T-loop phosphorylation.
Biorxiv, 2024
|
|
2ILA
 
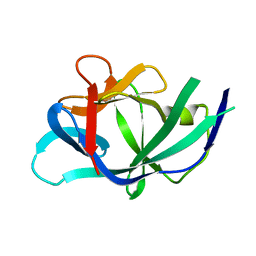 | |
7AXW
 
 | |
2XVW
 
 | |
7TM9
 
 | |
7QYS
 
 | | Crystal structure of RimK from Pseudomonas syringae DC3000 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase | | 著者 | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | 登録日 | 2022-01-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
7QHL
 
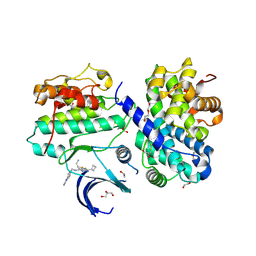 | | Crystal structure of Cyclin-dependent kinase 2/cyclin A in complex with 3,5,7-Substituted pyrazolo[4,3-d]pyrimidine inhibitor 24 | | 分子名称: | 1,2-ETHANEDIOL, 5-(2-amino-1-ethyl)thio-3-cyclobutyl-7-[4-(pyrazol-1-yl)benzyl]amino-1(2)H-pyrazolo[4,3-d]pyrimidine, Cyclin-A2, ... | | 著者 | Djukic, S, Skerlova, J, Rezacova, P. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 3,5,7-Substituted Pyrazolo[4,3- d ]Pyrimidine Inhibitors of Cyclin-Dependent Kinases and Cyclin K Degraders.
J.Med.Chem., 65, 2022
|
|
7ARC
 
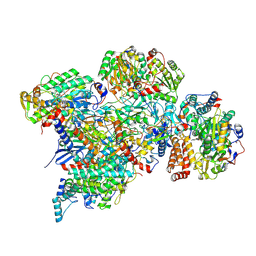 | | Cryo-EM structure of Polytomella Complex-I (peripheral arm) | | 分子名称: | 13 kDa, 18 kDa, 24 kDa, ... | | 著者 | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | 登録日 | 2020-10-23 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2N2X
 
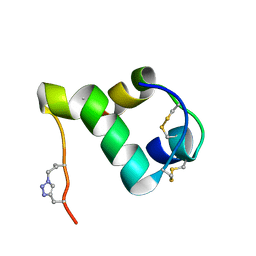 | |
7JN0
 
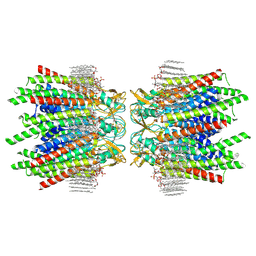 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-08-03 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2N52
 
 | |
7JPU
 
 | |
7QLP
 
 | | Structure of beta-lactamase TEM-171 complexed with tazobactam intermediate at 2.3 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Hakanpaa, J, Petrova, T, Samygina, V.R, Lamzin, V.S, Egorov, A.M. | | 登録日 | 2021-12-20 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of the molecular class A beta-lactamase TEM-171 and its complexes with tazobactam.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7JTS
 
 | | Stalk of radial spoke 1 attached with doublet microtubule from Chlamydomonas reinhardtii | | 分子名称: | Calmodulin, Dynein 8 kDa light chain, flagellar outer arm, ... | | 著者 | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | 登録日 | 2020-08-18 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARD
 
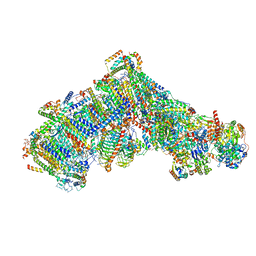 | | Cryo-EM structure of Polytomella Complex-I (complete composition) | | 分子名称: | (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 13 kDa, 15 kDa, ... | | 著者 | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | 登録日 | 2020-10-23 | | 公開日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2XXW
 
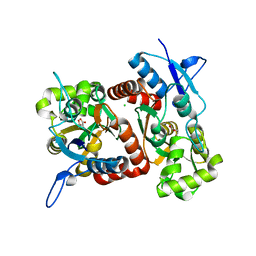 | | Crystal structure of the GluK2 (GluR6) D776K LBD dimer in complex with glutamate | | 分子名称: | CHLORIDE ION, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2010-11-12 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational Flexibility of the Ligand-Binding Domain Dimer in Kainate Receptor Gating and Desensitization
J.Neurosci., 31, 2011
|
|
7JVB
 
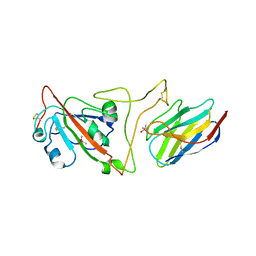 | | Crystal structure of the SARS-CoV-2 spike receptor-binding domain (RBD) with nanobody Nb20 | | 分子名称: | CACODYLATE ION, Nanobody Nb20, Spike protein S1 | | 著者 | Xiang, Y, Xiao, Z, Liu, H, Sang, Z, Schneidman-Duhovny, D, Zhang, C, Shi, Y. | | 登録日 | 2020-08-20 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.287 Å) | | 主引用文献 | Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science, 370, 2020
|
|
8PJA
 
 | | FKBP51FK1 F67E/K58 (i, i+9) in complex with SAFit1 | | 分子名称: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Meyners, C, Charalampidou, A, Hausch, F. | | 登録日 | 2023-06-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
2MP9
 
 | | Solution structure of an potent antifungal peptide Cm-p5 derived from C. muricatus | | 分子名称: | Antifungal peptide | | 著者 | Sun, Z.J, Heffron, G, Mcbeth, C, Wagner, G, Otero-Gonzales, A.J, Starnbach, M.N. | | 登録日 | 2014-05-14 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cm-p5: an antifungal hydrophilic peptide derived from the coastal mollusk Cenchritis muricatus (Gastropoda: Littorinidae).
Faseb J., 29, 2015
|
|
