8J91
 
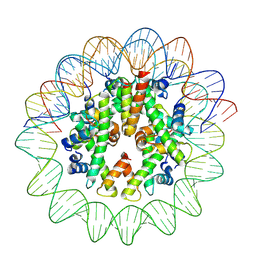 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | 分子名称: | DNA (169-MER), HTA13, Histone H2B.6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
3ISN
 
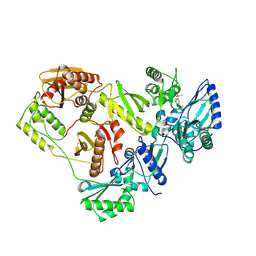 | | Crystal structure of HIV-1 RT bound to A 6-vinylpyrimidine inhibitor | | 分子名称: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Ennifar, E, Freisz, S, Bec, G, Dumas, P, Botta, M, Radi, M. | | 登録日 | 2009-08-26 | | 公開日 | 2010-03-16 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3ITH
 
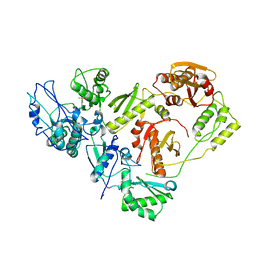 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | 分子名称: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | 登録日 | 2009-08-28 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8OTS
 
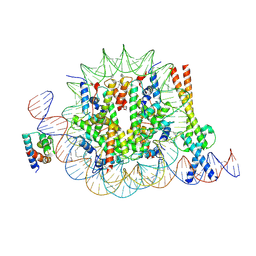 | | OCT4 and MYC-MAX co-bound to a nucleosome | | 分子名称: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
5KNR
 
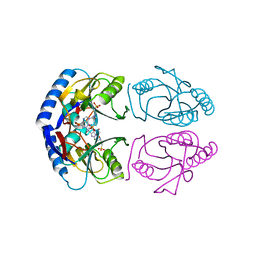 | | E. coli HPRT in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]-guanine | | 分子名称: | (2-{[2-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)ethyl][2-(2-phosphonoethoxy)ethyl]amino}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | 著者 | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | 登録日 | 2016-06-28 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.864 Å) | | 主引用文献 | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
6KXV
 
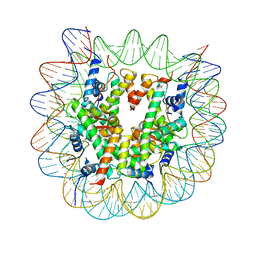 | | Crystal structure of a nucleosome containing Leishmania histone H3 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Dacher, M, Taguchi, H, Kujirai, T, Kurumizaka, H. | | 登録日 | 2019-09-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.63 Å) | | 主引用文献 | Incorporation and influence of Leishmania histone H3 in chromatin.
Nucleic Acids Res., 47, 2019
|
|
6T9L
 
 | |
9LIU
 
 | | Structure of isw1-nucleosome double-bound complex in ATP-ATP state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | 著者 | Sia, Y, Pan, H, Chen, Z. | | 登録日 | 2025-01-14 | | 公開日 | 2025-04-09 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
8UXQ
 
 | |
3LP1
 
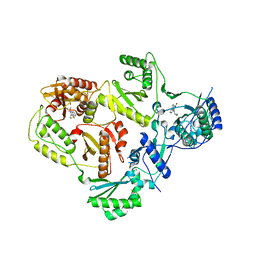 | | HIV-1 reverse transcriptase with inhibitor | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 3-cyclopentyl-1,4-dihydroxy-1,8-naphthyridin-2(1H)-one, MANGANESE (II) ION, ... | | 著者 | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | 登録日 | 2010-02-04 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP0
 
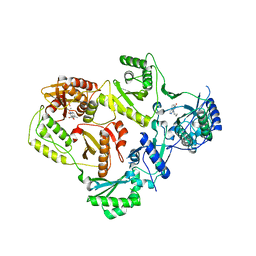 | | HIV-1 reverse transcriptase with inhibitor | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | 登録日 | 2010-02-04 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
4CD1
 
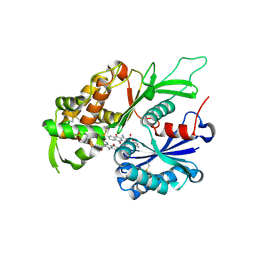 | | RnNTPDase2 in complex with PSB-071 | | 分子名称: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2 | | 著者 | Zebisch, M, Schaefer, P, Straeter, N. | | 登録日 | 2013-10-29 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
4CD3
 
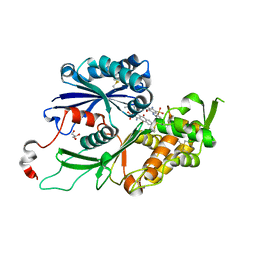 | | RnNTPDase2 X4 variant in complex with PSB-071 | | 分子名称: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | 著者 | Zebisch, M, Schaefer, P, Straeter, N. | | 登録日 | 2013-10-29 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
9JO5
 
 | | Structure of isw1-nucleosome complex in ADP-B state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | 著者 | Sia, Y, Pan, H, Chen, Z. | | 登録日 | 2024-09-24 | | 公開日 | 2025-04-09 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
1HQU
 
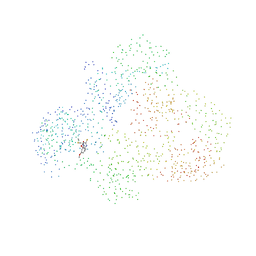 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | 分子名称: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | 著者 | Hsiou, Y, Ding, J, Arnold, E. | | 登録日 | 2000-12-19 | | 公開日 | 2001-05-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
5GPL
 
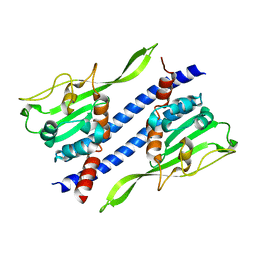 | | Crystal structure of Ccp1 | | 分子名称: | Putative nucleosome assembly protein C36B7.08c | | 著者 | Yin, F, Gao, F, Chen, Y. | | 登録日 | 2016-08-03 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
9LJ2
 
 | | Structure of isw1-nucleosome double-bound complex in ADP-ADP+ state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | 著者 | Sia, Y, Pan, H, Chen, Z. | | 登録日 | 2025-01-14 | | 公開日 | 2025-04-09 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
7M1X
 
 | |
8OSL
 
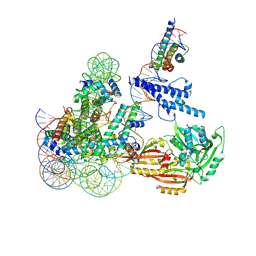 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | 分子名称: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | 著者 | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-04-19 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
4P54
 
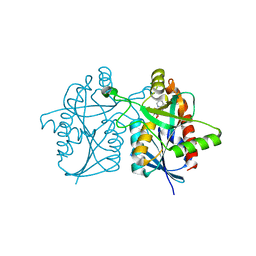 | |
7P0V
 
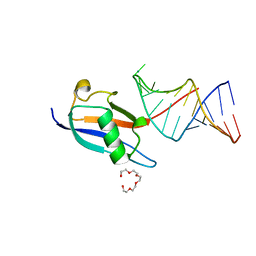 | | Crystal structure of human SF3A1 ubiquitin-like domain in complex with U1 snRNA stem-loop 4 | | 分子名称: | Isoform 2 of Splicing factor 3A subunit 1, PENTAETHYLENE GLYCOL, RNA (5'-R(P*GP*GP*GP*GP*AP*CP*UP*GP*CP*GP*UP*UP*CP*GP*CP*GP*CP*UP*UP*UP*CP*CP*CP*C)-3'), ... | | 著者 | Sabath, K, de Vries, T, Jonas, S. | | 登録日 | 2021-06-30 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3IS9
 
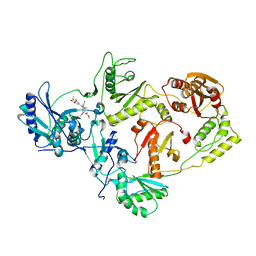 | | Crystal structure of the HIV-1 reverse transcriptase (RT) in complex with the alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate). | | 分子名称: | Reverse transcriptase, Reverse transcriptase/ribonuclease H, dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate) | | 著者 | Ho, W.C, Bauman, J.D, Das, K, Arnold, E. | | 登録日 | 2009-08-25 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystallographic study of a novel subnanomolar inhibitor provides insight on the binding interactions of alkenyldiarylmethanes with human immunodeficiency virus-1 reverse transcriptase.
J.Med.Chem., 52, 2009
|
|
7JHI
 
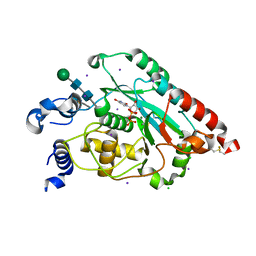 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 iodide-derivative | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Hao, Y, Huang, X. | | 登録日 | 2020-07-20 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
