7DYC
 
 | |
3HME
 
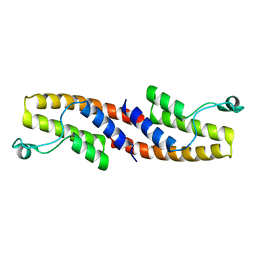 | | Crystal structure of human bromodomain containing 9 isoform 1 (BRD9) | | 分子名称: | Bromodomain-containing protein 9 | | 著者 | Filippakopoulos, P, Eswaran, J, Keates, T, Picaud, S, Roos, A, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-05-29 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7SN3
 
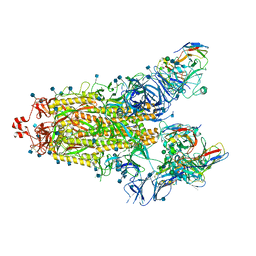 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Shankar, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
5L1H
 
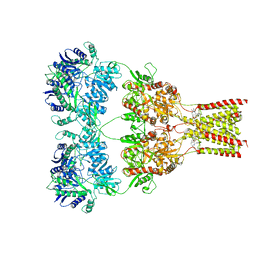 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | 分子名称: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | 著者 | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | 登録日 | 2016-07-29 | | 公開日 | 2016-10-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.801 Å) | | 主引用文献 | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
7APM
 
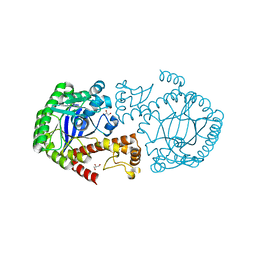 | | tRNA-guanine transglycosylase H319C mutant spin-labeled with MTSL. | | 分子名称: | CHLORIDE ION, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | 著者 | Nguyen, D, Heine, A, Klebe, G. | | 登録日 | 2020-10-18 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Unraveling a Ligand-Induced Twist of a Homodimeric Enzyme by Pulsed Electron-Electron Double Resonance.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DXV
 
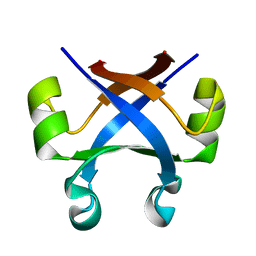 | |
2IH0
 
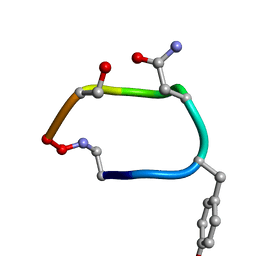 | |
7SN2
 
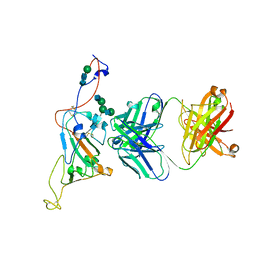 | | Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Yang, P, Shankar, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
2J4E
 
 | | THE ITP COMPLEX OF HUMAN INOSINE TRIPHOSPHATASE | | 分子名称: | INOSINE 5'-TRIPHOSPHATE, INOSINE TRIPHOSPHATE PYROPHOSPHATASE, INOSINIC ACID, ... | | 著者 | Stenmark, P, Kursula, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmbergschiavone, L, Hogbom, M, Kotenyova, T, Landry, R, Loppnau, P, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Schuler, H, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | 登録日 | 2006-08-29 | | 公開日 | 2006-09-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Human Inosine Triphosphatase: Substrate Binding and Implication of the Inosine Triphosphatase Deficiency Mutation P32T.
J.Biol.Chem., 282, 2007
|
|
7EK9
 
 | | Crystal structure of apo streptavidin at cryogenic temperature | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | 著者 | DeMirci, H, Ayan, E, Destan, E, Ertem, F.B. | | 登録日 | 2021-04-04 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Crystal structure of apo streptavidin at cryogenic temperature
To Be Published
|
|
2LX4
 
 | |
7SY9
 
 | |
7PGY
 
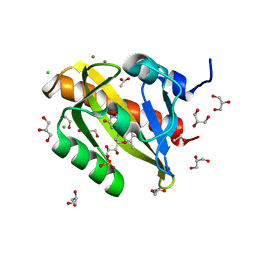 | | Structure of light-adapted AsLOV2 wild type | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Gelfert, R, Weyand, M, Moeglich, A. | | 登録日 | 2021-08-16 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
7K1D
 
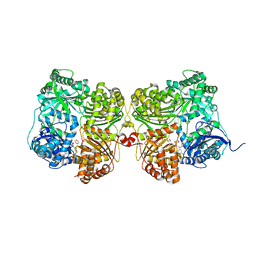 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | 分子名称: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
7DXU
 
 | |
7K1F
 
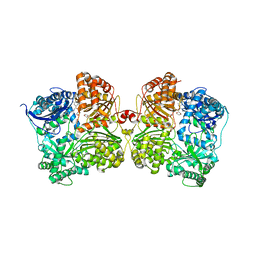 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88558 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-({1-[(2R)-4-(hydroxyamino)-4-oxo-1-(quinolin-7-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 4
To Be Published
|
|
7ARE
 
 | | DNA origami pointer object v2 | | 分子名称: | SCAFFOLD STRAND, STAPLE STRAND | | 著者 | Thomas, M, Feigl, E, Kohler, F, Kube, M, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | 登録日 | 2020-10-24 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
2LZZ
 
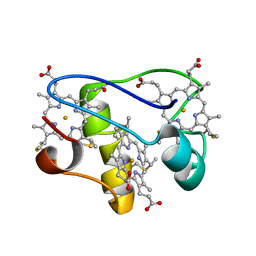 | | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15 | | 分子名称: | Cytochrome c, 3 heme-binding sites, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Dantas, J.M, Morgado, L, Turner, D.L, Salgueiro, C.A. | | 登録日 | 2012-10-12 | | 公開日 | 2013-01-30 | | 最終更新日 | 2013-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15.
Biochim.Biophys.Acta, 1827, 2013
|
|
2ZLE
 
 | |
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | 分子名称: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
2IJ5
 
 | |
7Q2O
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#1) | | 分子名称: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | 著者 | Loch, J.I, Barciszewski, J, Lewinski, K. | | 登録日 | 2021-10-25 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
2MC7
 
 | | Structure of Salmonella MgtR | | 分子名称: | Regulatory peptide | | 著者 | Jean-Francois, F, Dai, J, Yu, L, Myrick, A, Rubin, E, Fajer, P, Song, L, Zhou, H, Cross, T. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study.
J.Mol.Biol., 426, 2014
|
|
7K1E
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_88646 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 3,4-difluoro-N-[(1S)-1-{1-[(2R)-4-(hydroxyamino)-4-oxo-1-(5,6,7,8-tetrahydronaphthalen-2-yl)butan-2-yl]-1H-1,2,3-triazol-4-yl}ethyl]benzamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | 登録日 | 2020-09-07 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 3
To Be Published
|
|
7DXY
 
 | |
