6ZZX
 
 | | Structure of low-light grown Chlorella ohadii Photosystem I | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | 著者 | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | 登録日 | 2020-08-05 | | 公開日 | 2021-07-28 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
8CIC
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with clinical candidate Etrumadenant | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, ... | | 著者 | Cheng, R.K.Y, Markovic-Mueller, S, Hennig, M. | | 登録日 | 2023-02-09 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of adenosine A 2A receptor in complex with clinical candidate Etrumadenant reveals unprecedented antagonist interaction.
Commun Chem, 6, 2023
|
|
8JU5
 
 | | Structure of human TRPV4 with antagonist A1 | | 分子名称: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | 著者 | Fan, J, Lei, X. | | 登録日 | 2023-06-24 | | 公開日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 2024
|
|
4Q50
 
 | |
6EHP
 
 | |
6LQN
 
 | | EBV tegument protein BBRF2 | | 分子名称: | Cytoplasmic envelopment protein 1, NITRATE ION, SULFATE ION | | 著者 | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | 登録日 | 2020-01-14 | | 公開日 | 2020-10-07 | | 最終更新日 | 2021-04-21 | | 実験手法 | X-RAY DIFFRACTION (1.600022 Å) | | 主引用文献 | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
6XCT
 
 | |
6LQO
 
 | | EBV tegument protein BBRF2/BSRF1 complex | | 分子名称: | ACETATE ION, Cytoplasmic envelopment protein 1, GLYCEROL, ... | | 著者 | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | 登録日 | 2020-01-14 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.0911262 Å) | | 主引用文献 | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
6M32
 
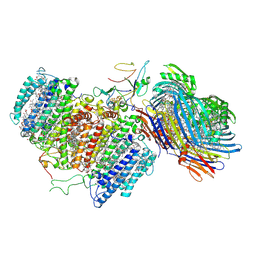 | | Cryo-EM structure of FMO-RC complex from green sulfur bacteria | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | 著者 | Chen, J.H, Zhang, X. | | 登録日 | 2020-03-02 | | 公開日 | 2020-11-25 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Architecture of the photosynthetic complex from a green sulfur bacterium.
Science, 370, 2020
|
|
1ARC
 
 | |
6LLF
 
 | | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase | | 分子名称: | 1,2-ETHANEDIOL, 3-(2-hydroxyphenyl)benzene-1,2-diol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | 登録日 | 2019-12-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Biphenyl-2,2',3-triol-soaked resting complex of Oxy and Fd in carbazole 1,9a-dioxygenase
To Be Published
|
|
7OYP
 
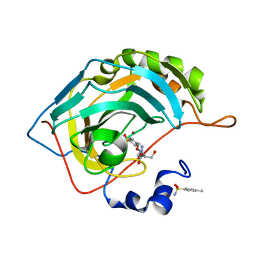 | | Carbonic anhydrase II in complex with Hit3-t1 (MH172) | | 分子名称: | (2S)-3-oxidanyl-2-[2-[(4-sulfamoylphenyl)methoxyamino]ethanoylamino]propanamide, 4-methylbenzenesulfonamide, Carbonic anhydrase 2, ... | | 著者 | Kugler, M, Brynda, J, Rezacova, P. | | 登録日 | 2021-06-24 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-29 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
7AW6
 
 | | The extracellular region of CD33 with bound sialoside analogue P22 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, ... | | 著者 | Bradshaw, W.J, Katis, V.L, Thompson, A.P, Arrowsmith, C.H, Edwards, A.M, von Delft, F, Bountra, C, Gileadi, O. | | 登録日 | 2020-11-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The extracellular region of CD33 with bound sialoside analogue P22
To Be Published
|
|
6ELO
 
 | |
1BAF
 
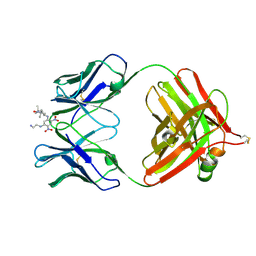 | | 2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN | | 分子名称: | IGG1-KAPPA AN02 FAB (HEAVY CHAIN), IGG1-KAPPA AN02 FAB (LIGHT CHAIN), N-(2-AMINO-ETHYL)-4,6-DINITRO-N'-(2,2,6,6-TETRAMETHYL-1-OXY-PIPERIDIN-4-YL)-BENZENE-1,3-DIAMINE | | 著者 | Leahy, D.J, Brunger, A.T, Fox, R.O, Hynes, T.R. | | 登録日 | 1992-01-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | 2.9 A resolution structure of an anti-dinitrophenyl-spin-label monoclonal antibody Fab fragment with bound hapten.
J.Mol.Biol., 221, 1991
|
|
6Y38
 
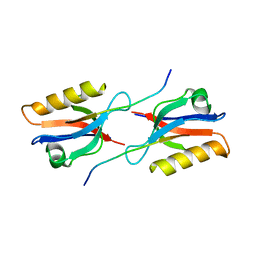 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | 分子名称: | Chains: C,D, Whirlin | | 著者 | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | 登録日 | 2020-02-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.697 Å) | | 主引用文献 | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6M8Q
 
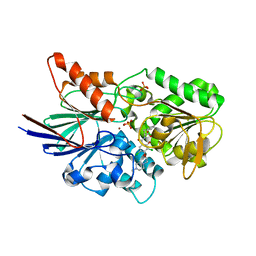 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | 分子名称: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | 著者 | Weihofen, W.A, Salcius, M, Michaud, G. | | 登録日 | 2018-08-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | 分子名称: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QKX
 
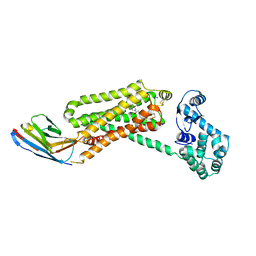 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | 分子名称: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | 著者 | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-23 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6YMX
 
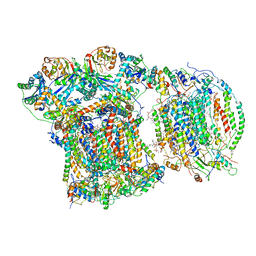 | | CIII2/CIV respiratory supercomplex from Saccharomyces cerevisiae | | 分子名称: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (1R)-2-(phosphonooxy)-1-[(tridecanoyloxy)methyl]ethyl pentadecanoate, (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, ... | | 著者 | Berndtsson, J, Rathore, S, Ott, M. | | 登録日 | 2020-04-10 | | 公開日 | 2020-09-09 | | 最終更新日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
6YID
 
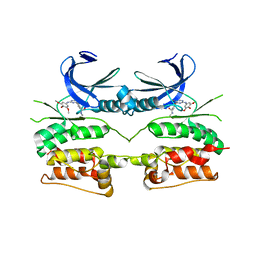 | | Crystal structure of ULK2 in complex with SBI-0206965 | | 分子名称: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, Serine/threonine-protein kinase ULK2 | | 著者 | Chaikuad, A, Ren, H, Bakas, N.A, Lambert, L.J, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-01 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design, Synthesis, and Characterization of an Orally Active Dual-Specific ULK1/2 Autophagy Inhibitor that Synergizes with the PARP Inhibitor Olaparib for the Treatment of Triple-Negative Breast Cancer.
J.Med.Chem., 63, 2020
|
|
4R4J
 
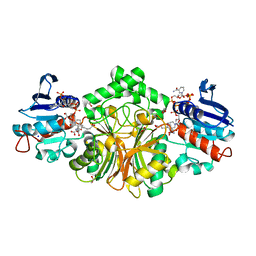 | | Crystal structure of complex sp_ASADH with 3-carboxypropyl-phthalic acid and Nicotinamide Adenine dinucleotide phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-(3-carboxypropyl)benzene-1,2-dicarboxylic acid, Aspartate-semialdehyde dehydrogenase, ... | | 著者 | Pavlovsky, A.G, Thangavelu, B, Bhansali, P, Viola, R.E. | | 登録日 | 2014-08-19 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | A cautionary tale of structure-guided inhibitor development against an essential enzyme in the aspartate-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W79
 
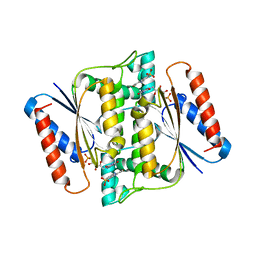 | | Crystal Structure of azoreductase AzrC in complex with sulfone-modified azo dye Orange I | | 分子名称: | 4-[(E)-(4-hydroxynaphthalen-1-yl)diazenyl]benzenesulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | 著者 | Ogata, D, Yu, J, Ooi, T, Yao, M. | | 登録日 | 2013-02-27 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P, Meents, A. | | 登録日 | 2020-10-21 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
