3L5B
 
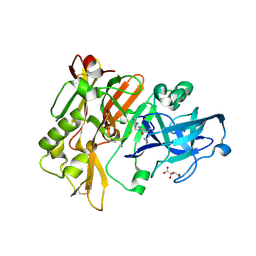 | | Structure of BACE Bound to SCH713601 | | 分子名称: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3MG6
 
 | | Structure of yeast 20S open-gate proteasome with Compound 6 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~2~-{[(1S)-6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl]acetyl}-N-{(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}-L-threoninamide, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2010-04-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3M88
 
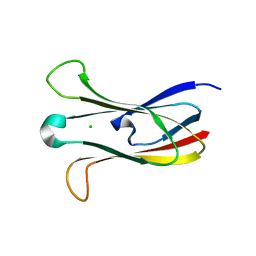 | | Crystal structure of the cysteine protease inhibitor, EhICP2, from Entamoeba histolytica | | 分子名称: | Amoebiasin-2, CHLORIDE ION, SULFATE ION | | 著者 | Lara-Gonzalez, S, Casados-Vazquez, L.E, Brieba, L.G. | | 登録日 | 2010-03-17 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the cysteine protease inhibitor 2 from Entamoeba histolytica: functional convergence of a common protein fold.
Gene, 471, 2011
|
|
4M9S
 
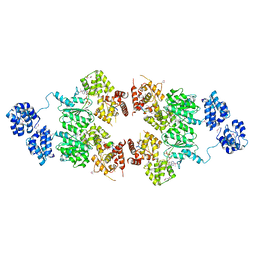 | | crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.207 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
3L5E
 
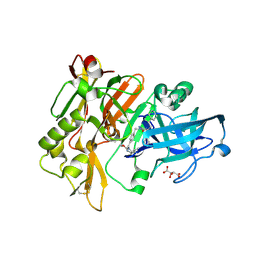 | | Structure of BACE Bound to SCH736062 | | 分子名称: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | 著者 | Strickland, C, Zhu, Z. | | 登録日 | 2009-12-21 | | 公開日 | 2010-02-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3MG0
 
 | | Structure of yeast 20S proteasome with bortezomib | | 分子名称: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, Proteasome component C1, Proteasome component C11, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2010-04-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3I25
 
 | | Potent Beta-Secretase 1 hydroxyethylene Inhibitor | | 分子名称: | Beta-secretase 1, N-[(2S,3S,5R)-1-(3,5-difluorophenoxy)-3-hydroxy-5-(2-methoxyethoxy)-6-[[(2S)-3-methyl-1-oxo-1-(phenylmethylamino)butan-2-yl]amino]-6-oxo-hexan-2-yl]-5-(methyl-methylsulfonyl-amino)-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | 著者 | Lindberg, J.D, Borkakoti, N, Nystrom, S. | | 登録日 | 2009-06-29 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of potent BACE-1 inhibitors containing a new hydroxyethylene (HE) scaffold: exploration of P1' alkoxy residues and an aminoethylene (AE) central core
Bioorg.Med.Chem., 18, 2010
|
|
4M9Z
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.405 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4U5Q
 
 | | High resolution crystal structure of reductase (R) domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Peptide synthetase | | 著者 | Patel, K.D, Haque, A.S, Priyadarshan, K, Sankaranarayanan, R. | | 登録日 | 2014-07-25 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.811 Å) | | 主引用文献 | High resolution structure of R-domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis
To Be Published
|
|
4JSU
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:3a | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | 著者 | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | 登録日 | 2013-03-22 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
3Q3X
 
 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | 分子名称: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | 著者 | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | 登録日 | 2010-12-22 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3MVI
 
 | | Crystal structure of holo mADA at 1.6 A resolution | | 分子名称: | Adenosine deaminase, GLYCEROL, ZINC ION | | 著者 | Niu, W, Shu, Q, Chen, Z, Mathews, S, Di Cera, E, Frieden, C. | | 登録日 | 2010-05-04 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The role of Zn2+ on the structure and stability of murine adenosine deaminase.
J.Phys.Chem.B, 114, 2010
|
|
3LC3
 
 | | Benzothiophene Inhibitors of Factor IXa | | 分子名称: | 1-[5-(3,4-dimethoxyphenyl)-1-benzothiophen-2-yl]methanediamine, CALCIUM ION, Coagulation factor IX | | 著者 | Wang, S, Beck, R. | | 登録日 | 2010-01-09 | | 公開日 | 2010-02-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Studies of Benzothiophene Template as Potent Factor IXa (FIXa) Inhibitors in Thrombosis.
J.Med.Chem., 53, 2010
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-07-02 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3Q45
 
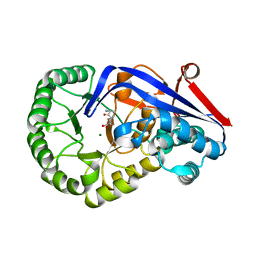 | | Crystal structure of Dipeptide Epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Val | | 分子名称: | D-ALANINE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme family; possible chloromuconate cycloisomerase, ... | | 著者 | Lukk, T, Gerlt, J.A, Nair, S.K. | | 登録日 | 2010-12-22 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KMX
 
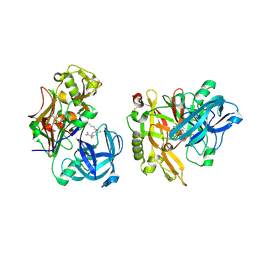 | | Structure of BACE bound to SCH346572 | | 分子名称: | 4-butoxy-3-chlorobenzyl imidothiocarbamate, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
4MVN
 
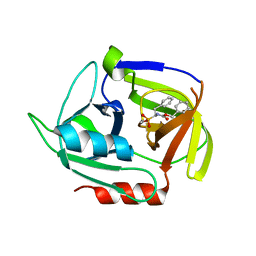 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | 分子名称: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | 著者 | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | 登録日 | 2013-09-24 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
3KYO
 
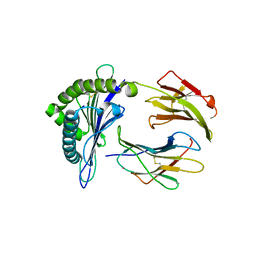 | | Crystal structure of HLA-G presenting KLPAQFYIL peptide | | 分子名称: | Beta-2-microglobulin, COBALT (II) ION, KLPAQFYIL peptide, ... | | 著者 | Walpole, N.G, Rossjohn, J, Clements, C.S. | | 登録日 | 2009-12-06 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structure and stability of the monomorphic HLA-G are influenced by the nature of the bound peptide
J.Mol.Biol., 397, 2010
|
|
4F49
 
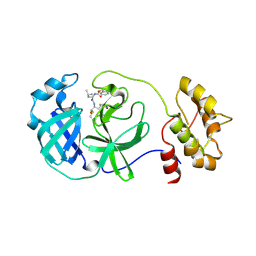 | | 2.25A resolution structure of Transmissible Gastroenteritis Virus Protease containing a covalently bound Dipeptidyl Inhibitor | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | 登録日 | 2012-05-10 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3Q3Y
 
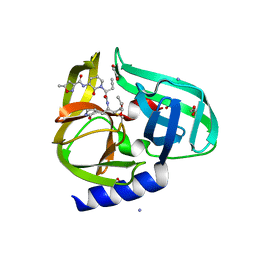 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | 分子名称: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | 著者 | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | 登録日 | 2010-12-22 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
2ZWL
 
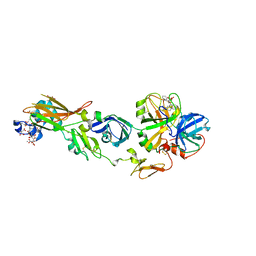 | | Human factor viia-tissue factor complexed with highly selective peptide inhibitor | | 分子名称: | 2-[[(2R)-1-[[(2S)-5-amino-1-[(4-carbamimidoylphenyl)methylamino]-1,5-dioxo-pentan-2-yl]amino]-3-(1H-indol-3-yl)-1-oxo-propan-2-yl]sulfamoyl]ethanoic acid, CALCIUM ION, Factor VII heavy chain, ... | | 著者 | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Sato, H, Ohta, M, Kozono, T. | | 登録日 | 2008-12-16 | | 公開日 | 2009-01-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Human factor viia-tissue factor complexed with highly selective peptide inhibitor
To be Published
|
|
4O9K
 
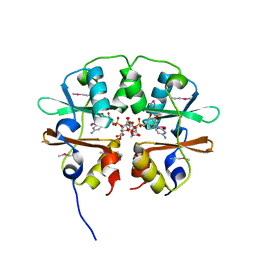 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | 分子名称: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | 著者 | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2014-01-02 | | 公開日 | 2014-01-22 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
3LIW
 
 | | Factor XA in complex with (R)-2-(1-ADAMANTYLCARBAMOYLAMINO)-3-(3-CARBAMIDOYL-PHENYL)-N-PHENETHYL-PROPIONIC ACID AMIDE | | 分子名称: | (R)-2-(3-ADAMANTAN-1-YL-UREIDO)-3-(3-CARBAMIMIDOYL-PHENYL)-N-PHENETHYL-PROPIONAMIDE, Activated factor Xa heavy chain, CALCIUM ION, ... | | 著者 | Mueller, M.M, Sperl, S, Sturzebecher, J, Bode, W, Moroder, L. | | 登録日 | 2010-01-25 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | (R)-3-Amidinophenylalanine-Derived Inhibitors of Factor Xa with a Novel Active-Site Binding Mode
Biol.Chem., 383, 2003
|
|
3KMY
 
 | | Structure of BACE bound to SCH12472 | | 分子名称: | 3-[2-(3-chlorophenyl)ethyl]pyridin-2-amine, Beta-secretase 1 | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
