1SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
1SGR
 
 | |
2AIP
 
 | |
1SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | 分子名称: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | 著者 | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | 登録日 | 1999-03-25 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1SGQ
 
 | |
1TAL
 
 | |
3B9F
 
 | | 1.6 A structure of the PCI-thrombin-heparin complex | | 分子名称: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, Plasma serine protease inhibitor, ... | | 著者 | Li, W, Adams, T.E, Huntington, J.A. | | 登録日 | 2007-11-05 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6CS2
 
 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
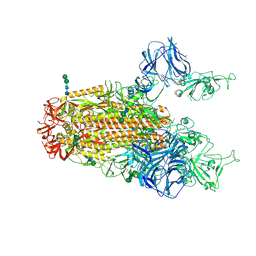 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRW
 
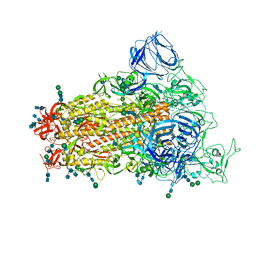 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRV
 
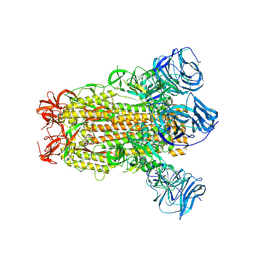 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
2D91
 
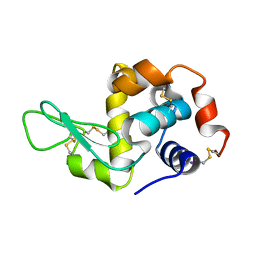 | | Structure of HYPER-VIL-lysozyme | | 分子名称: | IODIDE ION, Lysozyme C | | 著者 | Miyatake, H, Hasegawa, T, Yamano, A. | | 登録日 | 2005-12-08 | | 公開日 | 2006-07-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D97
 
 | | Structure of VIL-xylanase | | 分子名称: | Endo-1,4-beta-xylanase 2 | | 著者 | Miyatake, H, Hasegawa, T, Yamano, A. | | 登録日 | 2005-12-09 | | 公開日 | 2006-07-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
4DTG
 
 | | Hemostatic effect of a monoclonal antibody mAb 2021 blocking the interaction between FXa and TFPI in a rabbit hemophilia model | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Humanized recombinant FAB fragment, ... | | 著者 | Svensson, L.A, Breinholt, J, Krogh, B.O, Hilden, I. | | 登録日 | 2012-02-21 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hemostatic effect of a monoclonal antibody mAb 2021 blocking the interaction between FXa and TFPI in a rabbit hemophilia model.
Blood, 119, 2012
|
|
4DZR
 
 | | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-03-01 | | 公開日 | 2012-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
4M9I
 
 | |
4M9F
 
 | | Dengue virus NS2B-NS3 protease A125C variant at pH 8.5 | | 分子名称: | NS2B-NS3 protease | | 著者 | Yildiz, M, Ghosh, S, Bell, J.A, Sherman, W, Hardy, J.A. | | 登録日 | 2013-08-14 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus.
Acs Chem.Biol., 8, 2013
|
|
4M9K
 
 | |
4M9T
 
 | |
3TVJ
 
 | | Catalytic fragment of MASP-2 in complex with its specific inhibitor developed by directed evolution on SGCI scaffold | | 分子名称: | Mannan-binding lectin serine protease 2 A chain, Mannan-binding lectin serine protease 2 B chain, Protease inhibitor SGPI-2, ... | | 著者 | Heja, D, Harmat, V, Dobo, J, Szasz, R, Kekesi, K.A, Zavodszky, P, Gal, P, Pal, G. | | 登録日 | 2011-09-20 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Monospecific Inhibitors Show That Both Mannan-binding Lectin-associated Serine Protease-1 (MASP-1) and -2 Are Essential for Lectin Pathway Activation and Reveal Structural Plasticity of MASP-2.
J.Biol.Chem., 287, 2012
|
|
4M9M
 
 | |
6KK4
 
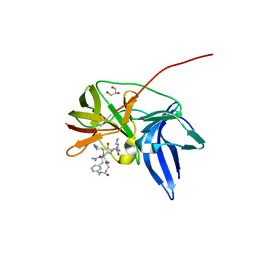 | | Crystal structure of Zika NS2B-NS3 protease with compound 9 | | 分子名称: | 1-[(9~{R},16~{S},19~{S})-16,19-bis(4-azanylbutyl)-4,8,15,18,21-pentakis(oxidanylidene)-3,7,14,17,20-pentazabicyclo[21.3.1]heptacosa-1(26),23(27),24-trien-9-yl]guanidine, GLYCEROL, NS3 protease, ... | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK5
 
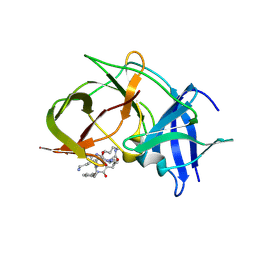 | | Crystal structure of Zika NS2B-NS3 protease with compound 15 | | 分子名称: | 1-[(5~{S},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KK2
 
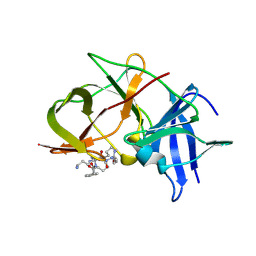 | | Crystal structure of Zika NS2B-NS3 protease with compound 2 | | 分子名称: | 1-[(10~{S},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6KPQ
 
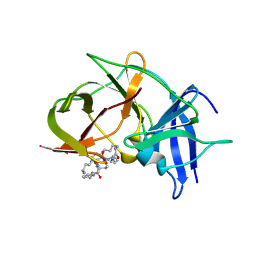 | | Crystal structure of Zika NS2B-NS3 protease with compound 8 | | 分子名称: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.3.1]octacosa-1(27),24(28),25-trien-10-yl]guanidine, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
