2VVR
 
 | |
3ZGL
 
 | | Crystal structures of Escherichia coli IspH in complex with AMBPP a potent inhibitor of the methylerythritol phosphate pathway | | 分子名称: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE, IRON/SULFUR CLUSTER | | 著者 | Borel, F, Barbier, E, Kratsutsky, S, Janthawornpong, K, Rohmer, M, Dale Poulter, C, Ferrer, J.L, Seemann, M. | | 登録日 | 2012-12-18 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Further Insight into Crystal Structures of Escherichia coli IspH/LytB in Complex with Two Potent Inhibitors of the MEP Pathway: A Starting Point for Rational Design of New Antimicrobials.
Chembiochem, 18, 2017
|
|
1K9S
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM E. COLI IN COMPLEX WITH FORMYCIN A DERIVATIVE AND PHOSPHATE | | 分子名称: | 2-(7-AMINO-6-METHYL-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 2-HYDROXYMETHYL-5-(7-METHYLAMINO-3H-PYRAZOLO[4,3-D]PYRIMIDIN-3-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, ... | | 著者 | Koellner, G, Bzowska, A, Wielgus-Kutrowska, B, Luic, M, Steiner, T, Saenger, W, Stepinski, J. | | 登録日 | 2001-10-30 | | 公開日 | 2001-11-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.
J.Mol.Biol., 315, 2002
|
|
1Q06
 
 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | 分子名称: | SILVER ION, Transcriptional regulator cueR | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
5MWQ
 
 | |
4XZJ
 
 | | Crystal structure of ADP-ribosyltransferase Vis in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative NAD(+)--arginine ADP-ribosyltransferase Vis | | 著者 | Pfoh, R, Ravulapalli, R, Merrill, A.R, Pai, E.F. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization of Vis Toxin, a Novel ADP-Ribosyltransferase from Vibrio splendidus.
Biochemistry, 54, 2015
|
|
6AZ3
 
 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | 分子名称: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | 著者 | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | 登録日 | 2017-09-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2022-04-13 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
4LF8
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | 分子名称: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | 著者 | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | 登録日 | 2013-06-26 | | 公開日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (3.1484 Å) | | 主引用文献 | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
6BTM
 
 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | 著者 | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | 登録日 | 2017-12-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
1I7W
 
 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | 著者 | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | 登録日 | 2020-12-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | 著者 | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | 登録日 | 2020-12-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
1FL3
 
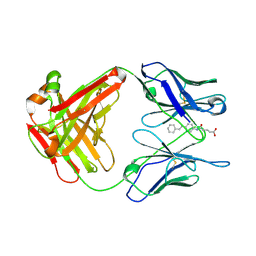 | |
6G7C
 
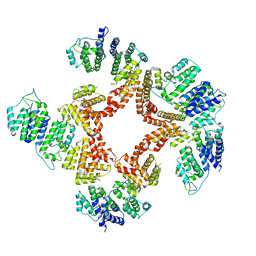 | | Nt2-CTD domains of the TssA component from the type VI secretion system of Aeromonas hydrophila. | | 分子名称: | ImpA-related domain protein | | 著者 | Dix, S.D, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | 登録日 | 2018-04-05 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
1KNK
 
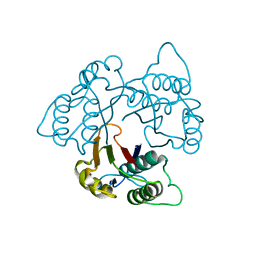 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | 分子名称: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | 著者 | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | 登録日 | 2001-12-18 | | 公開日 | 2002-06-18 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
7K54
 
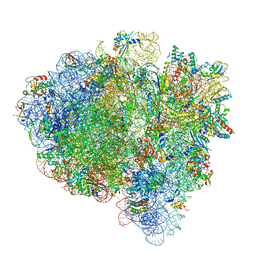 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K52
 
 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K53
 
 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
6ELZ
 
 | | State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | 著者 | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | 登録日 | 2017-09-30 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
6EOK
 
 | | Crystal structure of E. coli L-asparaginase II | | 分子名称: | L-asparaginase 2, ZINC ION | | 著者 | Cerofolini, L, Giuntini, S, Carlon, A, Ravera, E, Calderone, V, Fragai, M, Parigi, G, Luchinat, C. | | 登録日 | 2017-10-09 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Characterization of PEGylated Asparaginase: New Opportunities from NMR Analysis of Large PEGylated Therapeutics.
Chemistry, 25, 2019
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
5O4W
 
 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | 分子名称: | Lysozyme C | | 著者 | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | 主引用文献 | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6FM9
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | 分子名称: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | 著者 | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-01-30 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
