6HBA
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | 分子名称: | Carbon dioxide concentrating mechanism protein CcmM | | 著者 | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2018-08-10 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBC
 
 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | 分子名称: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | 著者 | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2018-08-10 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
4QFS
 
 | | Structure of AMPK in complex with Br2-A769662core activator and STAUROSPORINE inhibitor | | 分子名称: | 2-bromo-3-(4-bromophenyl)-4-hydroxy-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | 著者 | Calabrese, M.F, Kurumbail, R.G. | | 登録日 | 2014-05-21 | | 公開日 | 2014-08-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4RR2
 
 | | Crystal structure of human primase | | 分子名称: | DNA primase large subunit, DNA primase small subunit, IRON/SULFUR CLUSTER, ... | | 著者 | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | 登録日 | 2014-11-05 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of the human primase.
J.Biol.Chem., 290, 2015
|
|
6FVR
 
 | |
6IR9
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome | | 分子名称: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6INQ
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | 分子名称: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | 著者 | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | 登録日 | 2018-10-26 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6HBB
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | 分子名称: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | 著者 | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | 登録日 | 2018-08-10 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HEG
 
 | | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpB, PHOSPHATE ION, ... | | 著者 | Xin, B.G, Chen, W.F, Rety, S, Dai, Y.X, Xi, X.G. | | 登録日 | 2018-08-20 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.019 Å) | | 主引用文献 | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6IP6
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6MI0
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, ligand-free state | | 分子名称: | Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | 登録日 | 2018-09-18 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
5WGP
 
 | | Human Carbonic Anhydrase IX-mimic complexed with AceK | | 分子名称: | Acesulfame, Carbonic anhydrase 2, ZINC ION | | 著者 | Murray, A.B, Lomelino, C.L, Supuran, C.T, McKenna, R. | | 登録日 | 2017-07-14 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | "Seriously Sweet": Acesulfame K Exhibits Selective Inhibition Using Alternative Binding Modes in Carbonic Anhydrase Isoforms.
J. Med. Chem., 61, 2018
|
|
1MSH
 
 | |
5WEX
 
 | | Discovery of new selenoureido analogs of 4-(4-fluorophenylureido) benzenesulfonamides as carbonic anhydrase inhibitors | | 分子名称: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Angeli, A, Tanini, D, Peat, T.S, Di Cesare Mannelli, L, Bartolucci, G, Capperucci, A, Ghelardini, C, Supuran, C.T, Carta, F. | | 登録日 | 2017-07-10 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
1NTL
 
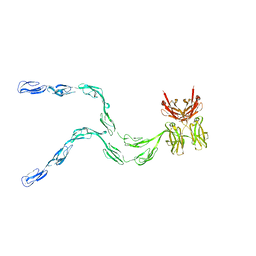 | | Model of mouse Crry-Ig determined by solution scattering, curve fitting and homology modelling | | 分子名称: | Complement component receptor 1-like protein,Ig gamma-1 chain C region secreted form | | 著者 | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | 登録日 | 2003-01-30 | | 公開日 | 2004-02-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | SOLUTION SCATTERING (30 Å) | | 主引用文献 | The extended multidomain solution structures of the complement protein Crry
and its chimaeric conjugate Crry-Ig by scattering, analytical ultracentrifugation
and constrained modelling: implications for function and therapy
J.Mol.Biol., 329, 2003
|
|
1NYN
 
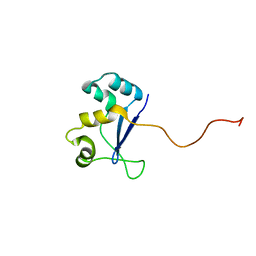 | | Solution NMR Structure of Protein YHR087W from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YTYST425. | | 分子名称: | Hypothetical 12.0 kDa protein in NAM8-GAR1 intergenic region | | 著者 | Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-02-13 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1O0D
 
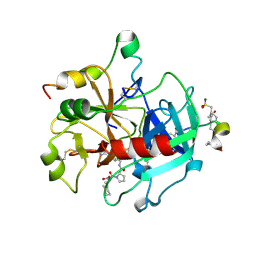 | | Human Thrombin complexed with a d-Phe-Pro-Arg-type Inhibitor and a C-terminal Hirudin derived exo-site inhibitor | | 分子名称: | (2-{2-[(5-CARBAMIMIDOYL-1-METHYL-1H-PYRROL-2-YLMETHYL)-CARBAMOYL]-PYRROL-1-YL}- 1-CYCLOHEXYLMETHYL-2-OXO-ETHYLAMINO)-ACETIC ACID, Decapeptide Hirudin Analogue, Thrombin heavy chain, ... | | 著者 | Lange, U.E, Bauke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | 登録日 | 2003-02-21 | | 公開日 | 2003-10-14 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | D-Phe-Pro-Arg type thrombin inhibitors: unexpected selectivity by modification of the P1 moiety
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2RMW
 
 | |
1NTJ
 
 | | Model of rat Crry determined by solution scattering, curve fitting and homology modelling | | 分子名称: | complement receptor related protein | | 著者 | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | 登録日 | 2003-01-30 | | 公開日 | 2004-02-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | SOLUTION SCATTERING (30 Å) | | 主引用文献 | The Extended Multidomain Solution Structures of the Complement Protein
Crry and its Chimeric Conjugate Crry-Ig by Scattering, Analytical
Ultracentrifugation and Constrained Modelling: Implications for Function and
Therapy
J.Mol.Biol., 329, 2003
|
|
4PAD
 
 | |
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | 分子名称: | Protein C10C5.1, isoform i | | 著者 | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | 登録日 | 2014-05-15 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
8IFD
 
 | | Dibekacin-added human 80S ribosome | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | 登録日 | 2023-02-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFE
 
 | | Arbekacin-added human 80S ribosome | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | 登録日 | 2023-02-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
5XEW
 
 | | Crystal structure of the [Ni2+-(chromomycin A3)2]-CCG repeats complex | | 分子名称: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | 著者 | Tseng, W.H, Wu, P.C, Hou, M.H. | | 登録日 | 2017-04-06 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel-Chromomycin Complex Resulting in Large-Scale DNA Deformation
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6ND1
 
 | | CryoEM structure of the Sec Complex from yeast | | 分子名称: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | 著者 | Wu, X, Cabanos, C, Rapoport, T.A. | | 登録日 | 2018-12-13 | | 公開日 | 2019-01-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of the post-translational protein translocation machinery of the ER membrane.
Nature, 566, 2019
|
|
