1EJY
 
 | |
1E4T
 
 | | Solution structure of the mouse defensin mBD-7 | | 分子名称: | Beta-defensin 7 | | 著者 | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | 登録日 | 2000-07-12 | | 公開日 | 2001-07-12 | | 最終更新日 | 2018-06-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
7T9W
 
 | | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2 | | 分子名称: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | 著者 | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-20 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2
To Be Published
|
|
1E4R
 
 | | Solution structure of the mouse defensin mBD-8 | | 分子名称: | Beta-defensin 8 | | 著者 | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | 登録日 | 2000-07-12 | | 公開日 | 2001-07-12 | | 最終更新日 | 2018-06-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1EJL
 
 | |
8B1L
 
 | |
3LG2
 
 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | 分子名称: | PHOSPHATE ION, Uncharacterized protein YKR043C | | 著者 | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-19 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
1E9W
 
 | | Structure of the macrocycle thiostrepton solved using the anomalous dispersive contribution from sulfur | | 分子名称: | THIOSTREPTON | | 著者 | Bond, C.S, Shaw, M.P, Alphey, M.S, Hunter, W.N. | | 登録日 | 2000-10-27 | | 公開日 | 2001-03-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Structure of the Macrocycle Thiostrepton Solved Using the Anomalous Dispersive Contribution from Sulfur
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7B7D
 
 | | Yeast 80S ribosome bound to eEF3 and A/A- and P/P-tRNAs | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Ranjan, N, Pochopien, A.A, Wu, C.C, Beckert, B, Blanchet, S, Green, R, Rodnina, M.V, Wilson, D.N. | | 登録日 | 2020-12-10 | | 公開日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | eEF3 promotes late stages of tRNA translocation including E-tRNA release from the ribosome
To Be Published
|
|
1ED0
 
 | | NMR structural determination of viscotoxin A3 from Viscum album L. | | 分子名称: | VISCOTOXIN A3 | | 著者 | Romagnoli, S, Ugolini, R, Fogolari, F, Schaller, G, Urech, K, Giannattasio, M, Ragona, L, Molinari, H. | | 登録日 | 2000-01-26 | | 公開日 | 2000-02-04 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural determination of viscotoxin A3 from Viscum album L.
Biochem.J., 350, 2000
|
|
6W5N
 
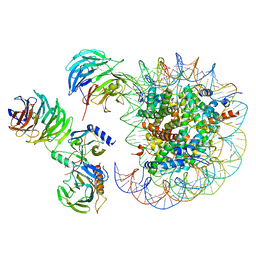 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | 分子名称: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | 登録日 | 2020-03-13 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5I
 
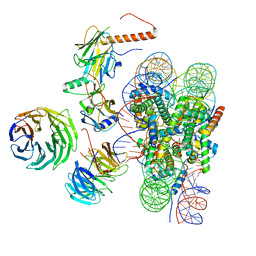 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | 分子名称: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | 登録日 | 2020-03-13 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
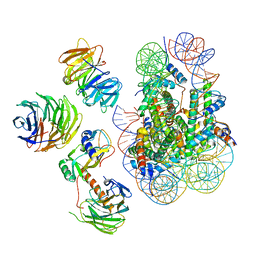 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | 分子名称: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | 登録日 | 2020-03-13 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-22 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
3LRU
 
 | |
4PKE
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | 分子名称: | PLATINUM (II) ION, Protein C10C5.1, isoform i | | 著者 | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | 登録日 | 2014-05-14 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
1EU1
 
 | | THE CRYSTAL STRUCTURE OF RHODOBACTER SPHAEROIDES DIMETHYLSULFOXIDE REDUCTASE REVEALS TWO DISTINCT MOLYBDENUM COORDINATION ENVIRONMENTS. | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | 著者 | Li, H.K, Temple, K, Rajagopalan, K.V, Schindelin, H. | | 登録日 | 2000-04-13 | | 公開日 | 2000-08-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The 1.3 A Crystal Structure of Rhodobacter sphaeroides Dimethylsulfoxide Reductase Reveals Two Distinct Molybdenum Coordination Environments
J.Am.Chem.Soc., 122, 2000
|
|
1E4S
 
 | | Solution structure of the human defensin hBD-1 | | 分子名称: | BETA-DEFENSIN 1 | | 著者 | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | 登録日 | 2000-07-12 | | 公開日 | 2001-07-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
6CGH
 
 | | Solution structure of the four-helix bundle region of human J-protein Zuotin, a component of ribosome-associated complex (RAC) | | 分子名称: | DnaJ homolog subfamily C member 2 | | 著者 | Shrestha, O.K, Lee, W, Tonelli, M, Cornilescu, G, Markley, J.L, Ciesielski, S.J, Craig, E.A. | | 登録日 | 2018-02-20 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70.
Plos One, 14, 2019
|
|
1EOH
 
 | | GLUTATHIONE TRANSFERASE P1-1 | | 分子名称: | GLUTATHIONE S-TRANSFERASE | | 著者 | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | 登録日 | 2000-03-22 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
7SPN
 
 | | Crystal structure of IS11, a thermophilic esterase | | 分子名称: | IS11 | | 著者 | Stogios, P.J, Evdokimova, E, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Crystal structure of IS11, a thermophilic esterase
To Be Published
|
|
7SNU
 
 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | 分子名称: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | 著者 | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | 登録日 | 2021-10-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
6ZMO
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S-eEF1a ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-03 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
4QR8
 
 | | Crystal Structure of E coli pepQ | | 分子名称: | MAGNESIUM ION, Xaa-Pro dipeptidase | | 著者 | Pingwei, L. | | 登録日 | 2014-06-30 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of substrate selectivity of E. coli prolidase.
Plos One, 9, 2014
|
|
5A0Q
 
 | |
