5FAK
 
 | | Crystal structure of Double Mutant S12T and N87T of Adenosine/Methylthioadenosine Phosphorylase from Schistosoma mansoni in complex with Adenine | | 分子名称: | ADENINE, Methylthioadenosine phosphorylase, SULFATE ION | | 著者 | Torini, J.R, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
8WUI
 
 | | SKOR D312N L271P double mutation | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Potassium channel SKOR | | 著者 | Gao, X, Sun, T, Lu, Y, Jia, Y, Xu, X, Zhang, Y, Fu, P, Yang, G. | | 登録日 | 2023-10-20 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural changes in the conversion of an Arabidopsis outward-rectifying K + channel into an inward-rectifying channel.
Plant Commun., 5, 2024
|
|
7JSG
 
 | | Adeno-Associated Virus 2 Rep68 HD-Heptamer-ssDNA with ATPgS | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | 著者 | Escalante, C.R. | | 登録日 | 2020-08-14 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
1N5W
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Oxidized form | | 分子名称: | CU(I)-S-MO(VI)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | 著者 | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | 登録日 | 2002-11-07 | | 公開日 | 2002-12-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7JY8
 
 | |
1N63
 
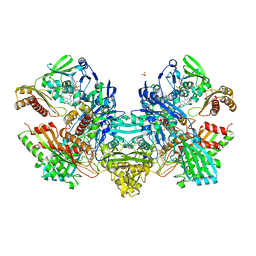 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Carbon monoxide reduced state | | 分子名称: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | 著者 | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | 登録日 | 2002-11-08 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4GX2
 
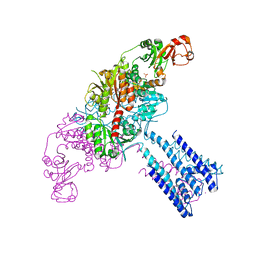 | | GsuK channel bound to NAD | | 分子名称: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, ... | | 著者 | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | 登録日 | 2012-09-03 | | 公開日 | 2012-12-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
1N61
 
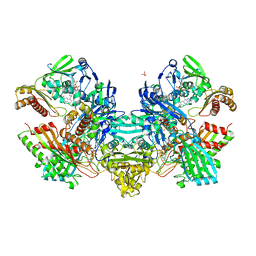 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Dithionite reduced state | | 分子名称: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | 著者 | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | 登録日 | 2002-11-08 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5D3W
 
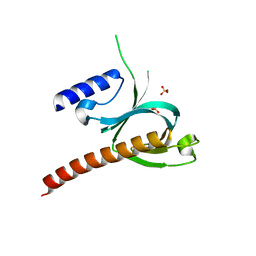 | | Crystal Structure of the P-Rex1 PH domain with Sulfate Bound | | 分子名称: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, SULFATE ION | | 著者 | Cash, J.N, Tesmer, J.J.G. | | 登録日 | 2015-08-06 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.852 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
8W20
 
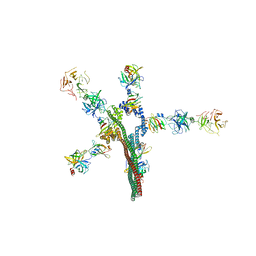 | | Umb1 umbrella toxin particle | | 分子名称: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | 著者 | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | 登録日 | 2024-02-19 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
1N6Z
 
 | |
5CRX
 
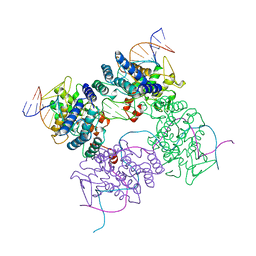 | |
8W09
 
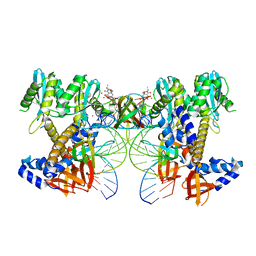 | | HIV-1 wild-type intasome core | | 分子名称: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, Integrase, MAGNESIUM ION, ... | | 著者 | Li, M, Craigie, R. | | 登録日 | 2024-02-13 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
5FCH
 
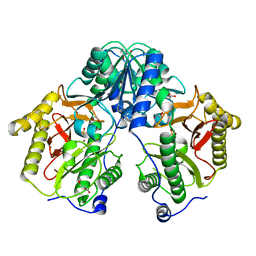 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | 著者 | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | 登録日 | 2015-12-15 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
8W2L
 
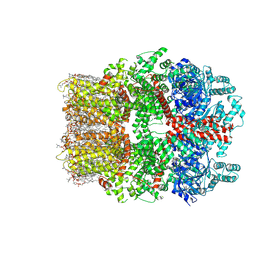 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | 著者 | Nadezhdin, K.D, Sobolevsky, A.I. | | 登録日 | 2024-02-20 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
2CD8
 
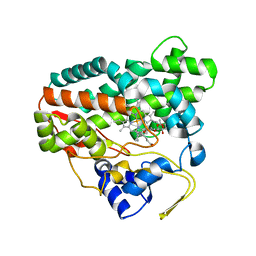 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | 分子名称: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | 登録日 | 2006-01-20 | | 公開日 | 2007-02-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
5FK3
 
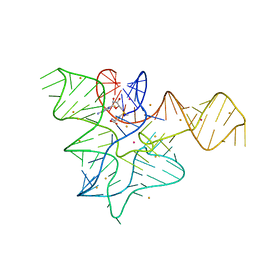 | |
5CSH
 
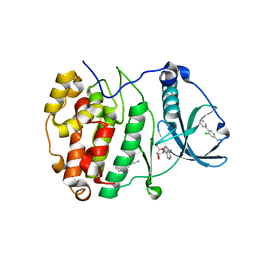 | | Crystal Structure of CK2alpha with Compound 4 bound | | 分子名称: | 1-(2-chlorobiphenyl-4-yl)methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | 登録日 | 2015-07-23 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8W34
 
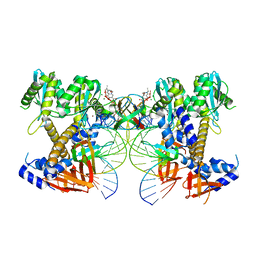 | | HIV-1 intasome core assembled with wild-type integrase, 1F | | 分子名称: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (5'-D(*AP*CP*TP*GP*CP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*CP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*GP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*GP*CP*A)-3'), ... | | 著者 | Li, M, Craigie, R. | | 登録日 | 2024-02-21 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.
J.Mol.Biol., 436, 2024
|
|
4GYN
 
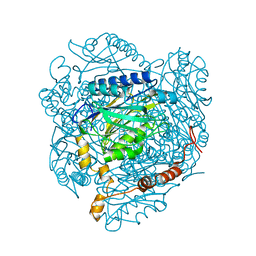 | | The E142L mutant of the amidase from Geobacillus pallidus | | 分子名称: | Aliphatic amidase, CHLORIDE ION | | 著者 | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | 登録日 | 2012-09-05 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
3FUA
 
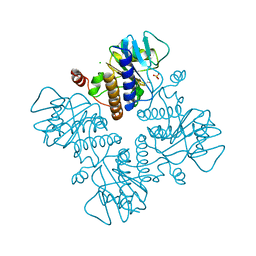 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE CRYSTAL FORM K | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | 著者 | Dreyer, M.K, Schulz, G.E. | | 登録日 | 1996-02-14 | | 公開日 | 1996-10-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
2VA5
 
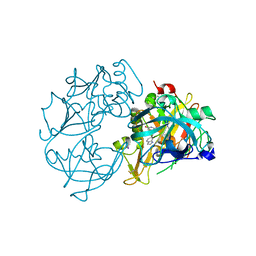 | | X-ray crystal structure of beta secretase complexed with compound 8c | | 分子名称: | 2-amino-6-[2-(1H-indol-6-yl)ethyl]pyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | 著者 | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L, Patel, S, Spear, N, Tian, G. | | 登録日 | 2007-08-30 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
5D4W
 
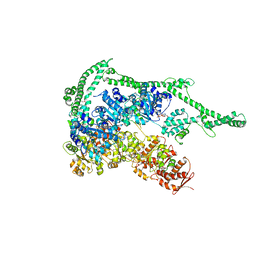 | |
2V1P
 
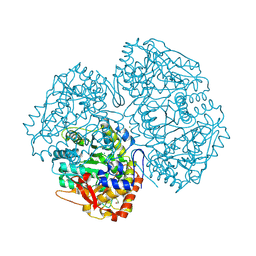 | | Crystal Structure of the apo form of Y74F mutant E. coli tryptophanase | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, TRYPTHOPANASE | | 著者 | Kogan, A, Gdalevsky, G.Y, Cohen-Luria, R, Goldgur, Y, Parola, A.H, Almog, O. | | 登録日 | 2007-05-28 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Changes and Loose Packing Promote E. Coli Tryptophanase Cold Lability.
Bmc Struct.Biol., 9, 2009
|
|
8W9B
 
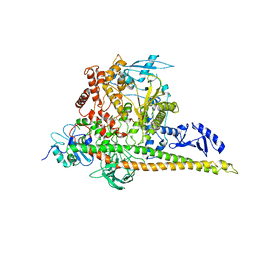 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | 分子名称: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Huang, X, Ren, X, Zhong, W. | | 登録日 | 2023-09-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
