3CZK
 
 | |
7NS3
 
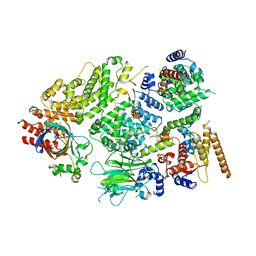 | | Substrate receptor scaffolding module of yeast Chelator-GID SR4 E3 ubiquitin ligase bound to Fbp1 substrate | | 分子名称: | BJ4_G0018240.mRNA.1.CDS.1, Fructose-bisphosphatase, Glucose-induced degradation protein 8, ... | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSB
 
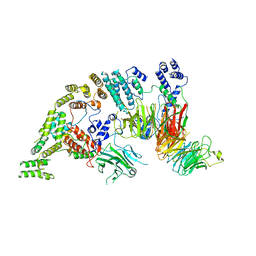 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | 分子名称: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NZ7
 
 | |
7NHN
 
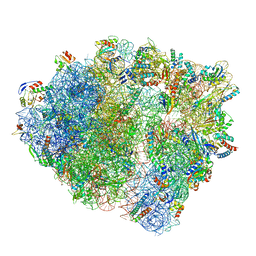 | | VgaL, an antibiotic resistance ABCF, in complex with 70S ribosome from Listeria monocytogenes | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Turnbull, K.J, Hauryliuk, V, Wilson, D.N. | | 登録日 | 2021-02-10 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NZ9
 
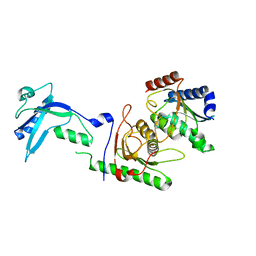 | |
7NHM
 
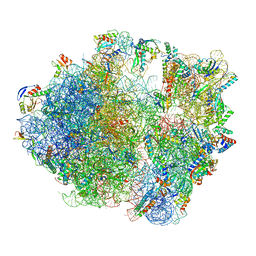 | | 70S ribosome from Staphylococcus aureus | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Crowe-McAuliffe, C, Murina, V, Hauryliuk, V, Wilson, D.N. | | 登録日 | 2021-02-10 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
3D12
 
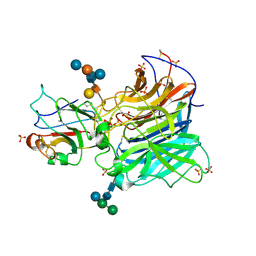 | | Crystal Structures of Nipah Virus G Attachment Glycoprotein in Complex with its Receptor Ephrin-B3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-B3, ... | | 著者 | Xu, K, Rajashankar, K.R, Chan, Y.P, Himanen, P, Broder, C.C, Nikolov, D.B. | | 登録日 | 2008-05-02 | | 公開日 | 2008-08-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7NRC
 
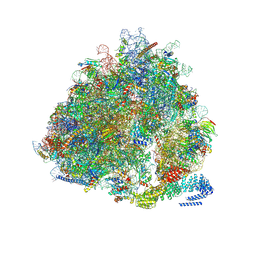 | | Structure of the yeast Gcn1 bound to a leading stalled 80S ribosome with Rbg2, Gir2, A- and P-tRNA and eIF5A | | 分子名称: | 18S rRNA (1771-MER), 25S rRNA (3184-MER), 40S ribosomal protein S0-A, ... | | 著者 | Pochopien, A.A, Beckert, B, Wilson, D.N. | | 登録日 | 2021-03-03 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NVG
 
 | | Salmonella flagellar basal body refined in C1 map | | 分子名称: | Basal-body rod modification protein FlgD, Flagellar L-ring protein, Flagellar M-ring protein, ... | | 著者 | Johnson, S, Furlong, E, Lea, S.M. | | 登録日 | 2021-03-15 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7NHK
 
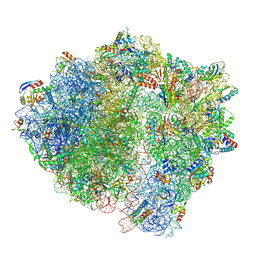 | | LsaA, an antibiotic resistance ABCF, in complex with 70S ribosome from Enterococcus faecalis | | 分子名称: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | 著者 | Crowe-McAuliffe, C, Kasari, M, Hauryliuk, V.H, Wilson, D.N. | | 登録日 | 2021-02-10 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NSC
 
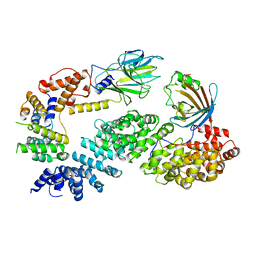 | | Substrate receptor scaffolding module of human CTLH E3 ubiquitin ligase | | 分子名称: | Glucose-induced degradation protein 4 homolog, Glucose-induced degradation protein 8 homolog, Isoform 2 of Armadillo repeat-containing protein 8, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NQL
 
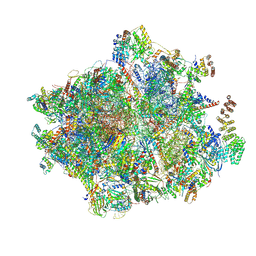 | | 55S mammalian mitochondrial ribosome with ICT1 and P site tRNAMet | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | 著者 | Kummer, E, Schubert, K, Ban, N. | | 登録日 | 2021-03-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NZ8
 
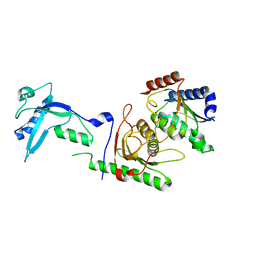 | |
7NS4
 
 | | Catalytic module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | 分子名称: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, ZINC ION | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NQH
 
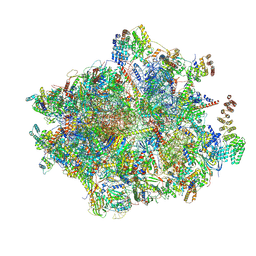 | | 55S mammalian mitochondrial ribosome with mtRF1a and P-site tRNAMet | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | 著者 | Kummer, E, Schubert, K, Ban, N. | | 登録日 | 2021-03-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NSH
 
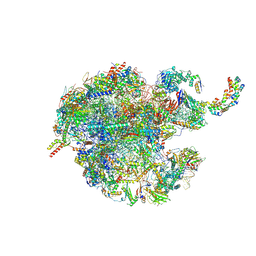 | | 39S mammalian mitochondrial large ribosomal subunit with mtRRF (post) and mtEFG2 | | 分子名称: | 16S rRNA, 39S ribosomal protein L48, mitochondrial, ... | | 著者 | Kummer, E, Schubert, K, Ban, N. | | 登録日 | 2021-03-07 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NXJ
 
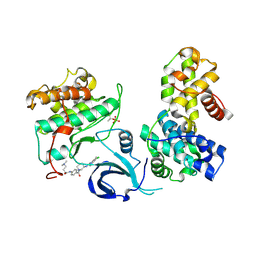 | | Crystal structure of human Cdk13/Cyclin K in complex with the inhibitor THZ531 | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 13, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | 著者 | Anand, K, Greifenberg, A.K, Kaltheuner, I.H, Geyer, M. | | 登録日 | 2021-03-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
7NXK
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with the inhibitor BSJ-01-175 | | 分子名称: | (E)-N-[4-[(1R,3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]cyclohexyl]oxyphenyl]-4-(dimethylamino)but-2-enamide, Cyclin-K, Cyclin-dependent kinase 12 | | 著者 | Anand, K, Dust, S, Kaltheuner, I.H, Geyer, M. | | 登録日 | 2021-03-18 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
7O56
 
 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element solved by Phosphorus and Sulphur SAD methods | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*AP*GP*AP*AP*AP*CP*CP*GP*AP*AP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*TP*TP*CP*GP*GP*TP*TP*TP*CP*TP*TP*TP*TP*AP*T)-3'), Interferon regulatory factor 4 | | 著者 | El Omari, K, Agnarelli, A, Duman, R, Wagner, A, Mancini, E.J. | | 登録日 | 2021-04-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7NUM
 
 | |
7O1H
 
 | |
7NUO
 
 | | Rhinovirus 14 empty particle at pH 6.2 | | 分子名称: | Genome polyprotein, P1 | | 著者 | Hrebik, D, Plevka, P. | | 登録日 | 2021-03-12 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NSY
 
 | |
7NSZ
 
 | | Drosophila PGRP-LB Y78F mutant | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Isoform A of Peptidoglycan-recognition protein LB, SODIUM ION, ... | | 著者 | Orlans, J, Aller, P, Da Silva, P. | | 登録日 | 2021-03-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | PGRP-LB: An Inside View into the Mechanism of the Amidase Reaction.
Int J Mol Sci, 22, 2021
|
|
