1IAR
 
 | | INTERLEUKIN-4 / RECEPTOR ALPHA CHAIN COMPLEX | | 分子名称: | PROTEIN (INTERLEUKIN-4 RECEPTOR ALPHA CHAIN), PROTEIN (INTERLEUKIN-4) | | 著者 | Hage, T, Sebald, W, Reinemer, P. | | 登録日 | 1999-02-25 | | 公開日 | 2000-03-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface.
Cell(Cambridge,Mass.), 97, 1999
|
|
1UO4
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | 分子名称: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | 著者 | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | 登録日 | 2003-09-15 | | 公開日 | 2004-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
4RUJ
 
 | | Crystal structure of zVDR L337H mutant-VD complex | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Huet, T, Moras, D, Rochel, N. | | 登録日 | 2014-11-20 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.352 Å) | | 主引用文献 | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
4S2Z
 
 | |
1HIJ
 
 | |
1PP8
 
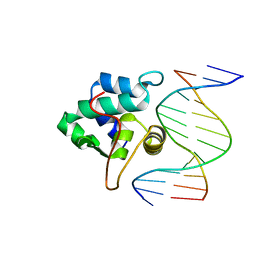 | | crystal structure of the T. vaginalis IBP39 Initiator binding domain (IBD) bound to the alpha-SCS Inr element | | 分子名称: | 39 kDa initiator binding protein, ALPHA-SCS INR, SULFATE ION | | 著者 | Schumacher, M.A, Lau, A.O.T, Johnson, P.J. | | 登録日 | 2003-06-16 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural Basis of Core Promoter Recognition in a Primitive Eukaryote
Cell(Cambridge,Mass.), 115, 2003
|
|
1HIK
 
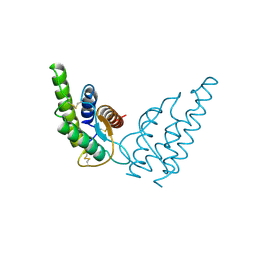 | | INTERLEUKIN-4 (WILD-TYPE) | | 分子名称: | INTERLEUKIN-4 | | 著者 | Mueller, T, Buehner, M. | | 登録日 | 1995-06-01 | | 公開日 | 1996-01-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models.
J.Mol.Biol., 247, 1995
|
|
2COB
 
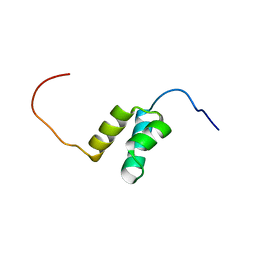 | | Solution structures of the HTH domain of human LCoR protein | | 分子名称: | LCoR protein | | 著者 | Nameki, N, Umehara, T, Sato, M, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-17 | | 公開日 | 2005-11-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of the HTH domain of human LCoR protein
To be Published
|
|
1ESX
 
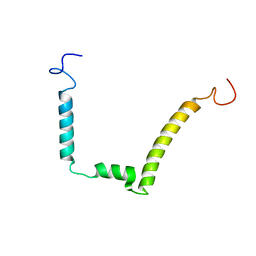 | | 1H, 15N AND 13C STRUCTURE OF THE HIV-1 REGULATORY PROTEIN VPR : COMPARISON WITH THE N-AND C-TERMINAL DOMAIN STRUCTURE, (1-51)VPR AND (52-96)VPR | | 分子名称: | VPR PROTEIN | | 著者 | Wecker, K, Morellet, N, Bouaziz, S, Roques, B. | | 登録日 | 2000-04-11 | | 公開日 | 2001-04-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the HIV-1 regulatory protein Vpr in H2O/trifluoroethanol. Comparison with the Vpr N-terminal (1-51) and C-terminal (52-96) domains.
Eur.J.Biochem., 269, 2002
|
|
1HZI
 
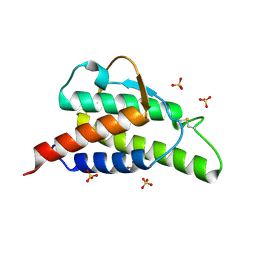 | | INTERLEUKIN-4 MUTANT E9A | | 分子名称: | INTERLEUKIN-4, SULFATE ION | | 著者 | Hulsmeyer, M, Scheufler, C, Dreyer, M.K. | | 登録日 | 2001-01-25 | | 公開日 | 2001-08-29 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of interleukin 4 mutant E9A suggests polar steering in receptor-complex formation.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2LGC
 
 | |
2JWO
 
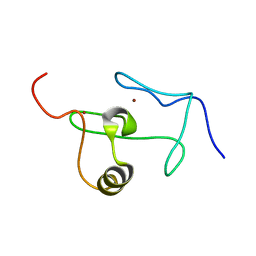 | | A PHD finger motif in the C-terminus of RAG2 modulates recombination activity | | 分子名称: | V(D)J recombination-activating protein 2, ZINC ION | | 著者 | Ivanov, D, Hyberts, S.G, Sun, Z, Wagner, G. | | 登録日 | 2007-10-17 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A PHD finger motif in the C terminus of RAG2 modulates recombination activity.
J.Biol.Chem., 280, 2005
|
|
7KKQ
 
 | |
4WF9
 
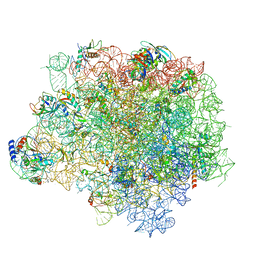 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | 登録日 | 2014-09-14 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.427 Å) | | 主引用文献 | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7ZUB
 
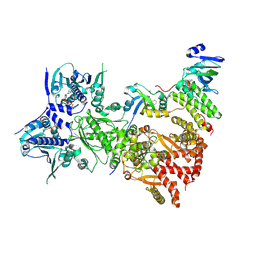 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | 分子名称: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | 著者 | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | 分子名称: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | 著者 | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | 登録日 | 2014-10-01 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
4WQK
 
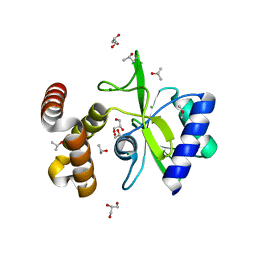 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | 分子名称: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.482 Å) | | 主引用文献 | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WTH
 
 | |
3VA2
 
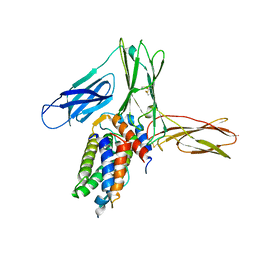 | | Crystal structure of human Interleukin-5 in complex with its alpha receptor | | 分子名称: | Interleukin-5, Interleukin-5 receptor subunit alpha | | 著者 | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | 登録日 | 2011-12-28 | | 公開日 | 2012-07-25 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Structural basis of interleukin-5 dimer recognition by its alpha receptor
Protein Sci., 21, 2012
|
|
7KRE
 
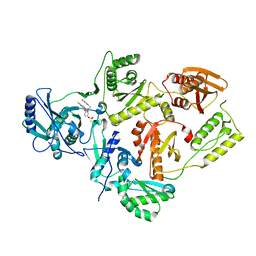 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-((6-cyanonaphthalen-1-yl)oxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ704) | | 分子名称: | 4-[(6-cyanonaphthalen-1-yl)oxy]-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | 著者 | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-11-19 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.728 Å) | | 主引用文献 | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
7KKP
 
 | |
4WMW
 
 | |
4WQL
 
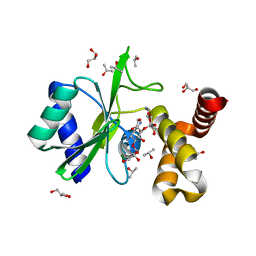 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, kanamycin-bound | | 分子名称: | 2''-aminoglycoside nucleotidyltransferase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WVI
 
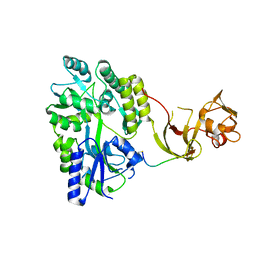 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep2). | | 分子名称: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep2) | | 著者 | Young, P.G, Ting, Y.T, Baker, E.N. | | 登録日 | 2014-11-05 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
4WVF
 
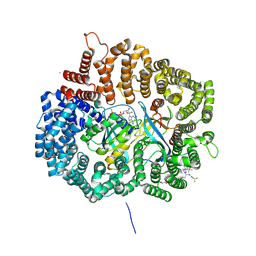 | | Crystal structure of KPT276 in complex with CRM1-Ran-RanBP1 | | 分子名称: | (2E)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-1-(3,3-difluoroazetidin-1-yl)prop-2-en-1-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Sun, Q, Chook, Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination.
Nat.Neurosci., 18, 2015
|
|
