8GTQ
 
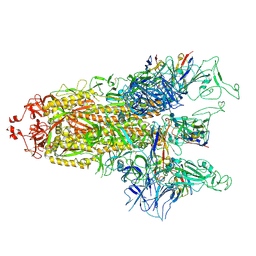 | | cryo-EM structure of Omicron BA.5 S protein in complex with S2L20 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | 登録日 | 2022-09-08 | | 公開日 | 2023-07-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
7QSO
 
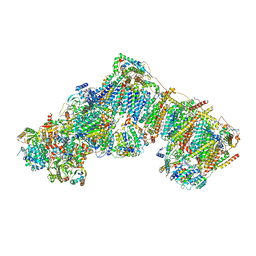 | | Bovine complex I in lipid nanodisc, State 3 (Slack) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
8DM5
 
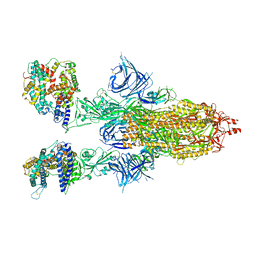 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
7QUS
 
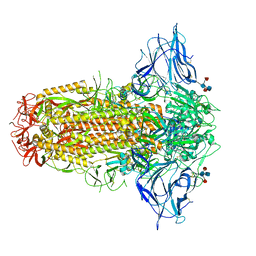 | | SARS-CoV-2 Spike, C3 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Naismith, J.H, Yang, Y, Liu, J.W. | | 登録日 | 2022-01-18 | | 公開日 | 2022-06-08 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
1DUL
 
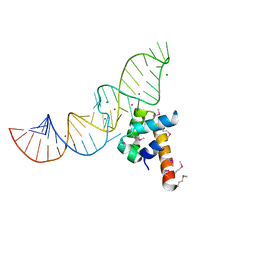 | | STRUCTURE OF THE RIBONUCLEOPROTEIN CORE OF THE E. COLI SIGNAL RECOGNITION PARTICLE | | 分子名称: | 4.5 S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Batey, R.T, Rambo, R.P, Lucast, L, Rha, B, Doudna, J.A. | | 登録日 | 2000-01-17 | | 公開日 | 2000-02-28 | | 最終更新日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the ribonucleoprotein core of the signal recognition particle.
Science, 287, 2000
|
|
7QIX
 
 | | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | | 分子名称: | 18S rRNA body, 30S ribosomal protein S15, chloroplastic, ... | | 著者 | Cottilli, P, Itoh, Y, Amunts, A. | | 登録日 | 2021-12-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
8SCB
 
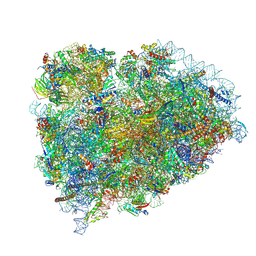 | | Terminating ribosome with SRI-41315 | | 分子名称: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | 著者 | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
8GK3
 
 | |
5CQ0
 
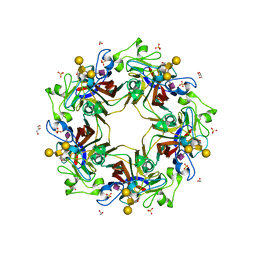 | | Crystal structure of murine polyomavirus RA strain VP1 in complex with the GD1a glycan | | 分子名称: | Capsid protein VP1, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | 著者 | Buch, M.H.C, Liaci, A.M, Neu, U, Stehle, T. | | 登録日 | 2015-07-21 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity.
Plos Pathog., 11, 2015
|
|
8SPE
 
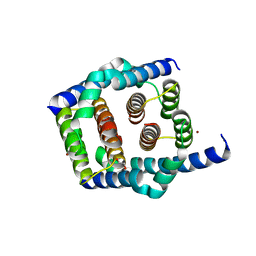 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | 分子名称: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2023-05-03 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
7QIY
 
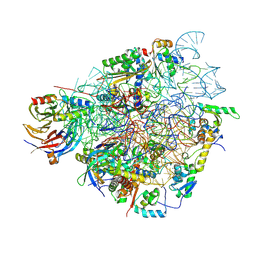 | | Specific features and methylation sites of a plant ribosome. 40S head ribosomal subunit. | | 分子名称: | 1,4-DIAMINOBUTANE, 18S rRNA head, 40S head ribosomal protein eS19, ... | | 著者 | Cottilli, P, Itoh, Y, Amunts, A. | | 登録日 | 2021-12-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7Q7Q
 
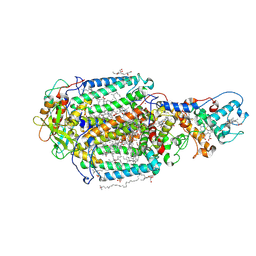 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | 著者 | Baath, P, Banacore, A, Neutze, R. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q7P
 
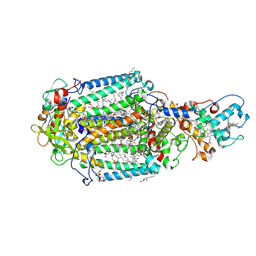 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | 著者 | Baath, P, Banacore, A, Neutze, R. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8G9V
 
 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | 分子名称: | 17-beta-hydroxysteroid dehydrogenase 13, 4-{[2,5-dimethyl-3-(4-methylbenzene-1-sulfonyl)benzene-1-sulfonyl]amino}benzoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Liu, S. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.645 Å) | | 主引用文献 | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
7QGR
 
 | | Structure of the SmrB-bound E. coli disome - collided 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | 登録日 | 2021-12-09 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
8SH3
 
 | | Pendrin in complex with iodide | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, IODIDE ION, ... | | 著者 | Wang, L, Hoang, A, Zhou, M. | | 登録日 | 2023-04-13 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
1DVL
 
 | |
6MSB
 
 | |
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | 著者 | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-10-01 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7QQH
 
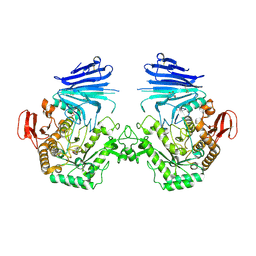 | | Crystal structure of MYORG (D520N) in complex with Gal-a1,4-Glc | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Meek, R.W, Davies, G.J. | | 登録日 | 2022-01-07 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
8SGW
 
 | | Pendrin in complex with chloride | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Wang, L, Hoang, A, Zhou, M. | | 登録日 | 2023-04-13 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SHC
 
 | | Pendrin in complex with Niflumic acid | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, ... | | 著者 | Wang, L, Hoang, A, Zhou, M. | | 登録日 | 2023-04-13 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SIE
 
 | | Pendrin in complex with bicarbonate | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, BICARBONATE ION, CHOLESTEROL, ... | | 著者 | Wang, L, Hoang, A, Zhou, M. | | 登録日 | 2023-04-16 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
7Q6I
 
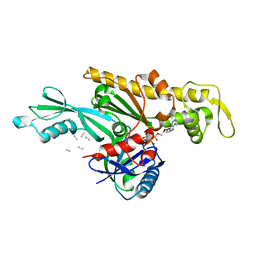 | | Vibrio maritimus FtsA 1-396 ATP and FtsN 1-29, bent tetramers in double filament arrangement | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division protein FtsA, Cell division protein FtsN (polyAla model), ... | | 著者 | Nierhaus, T, Kureisaite-Ciziene, D, Lowe, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol, 7, 2022
|
|
7QV7
 
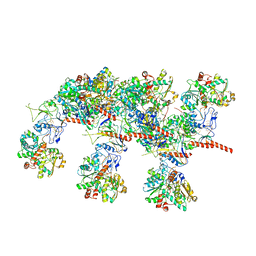 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | 分子名称: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | 著者 | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | 登録日 | 2022-01-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
