5MWW
 
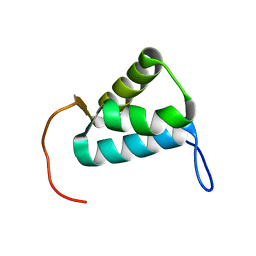 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | 分子名称: | RNA polymerase sigma factor SigA | | 著者 | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | 登録日 | 2017-01-20 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
2K6X
 
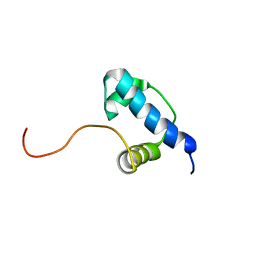 | | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1- Induced Compacted Structure | | 分子名称: | RNA polymerase sigma factor rpoD | | 著者 | Schwartz, E.C, Shekhtman, A, Dutta, K, Pratt, M.R, Cowburn, D, Darst, S, Muir, T.W. | | 登録日 | 2008-07-28 | | 公開日 | 2008-10-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1 - Induced Compacted Structure
Chem.Biol., 15, 2008
|
|
8Y6U
 
 | |
5VSW
 
 | |
6LDI
 
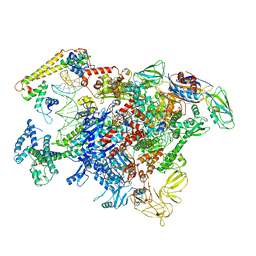 | |
8FTD
 
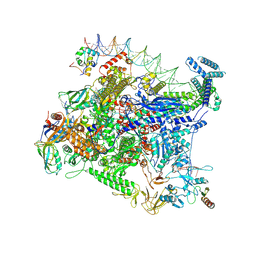 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | 分子名称: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Liu, M, Vassyliev, N, Nudler, E. | | 登録日 | 2023-01-11 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | 著者 | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | 登録日 | 2009-08-01 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (19.799999 Å) | | 主引用文献 | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8AD1
 
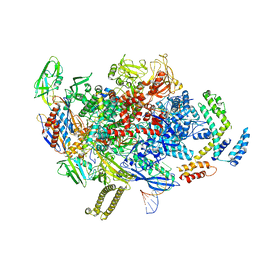 | |
7C17
 
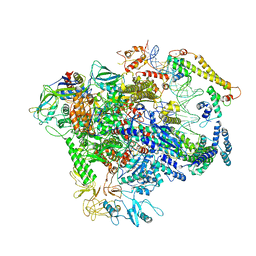 | |
5W1T
 
 | |
8U3B
 
 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | 分子名称: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Kompaniiets, D, Wang, D. | | 登録日 | 2023-09-07 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
6VJS
 
 | |
6JNX
 
 | | Cryo-EM structure of a Q-engaged arrested complex | | 分子名称: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Feng, Y, Shi, J. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.08 Å) | | 主引用文献 | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
7SZK
 
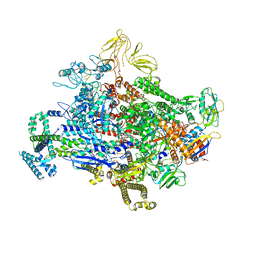 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | 分子名称: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Shin, Y, Murakami, K.S. | | 登録日 | 2021-11-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
8X6F
 
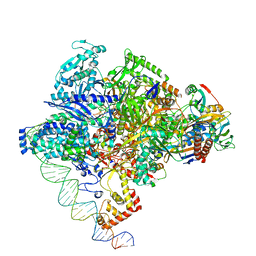 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | 分子名称: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | 登録日 | 2023-11-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
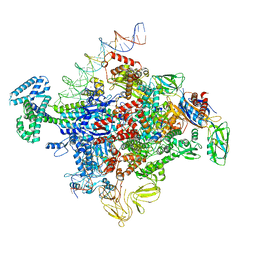
7SZJ
 
 | |
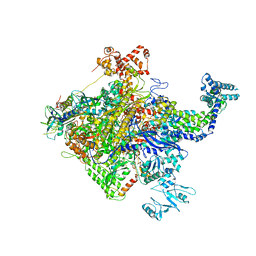
5W1S
 
 | |
6WMT
 
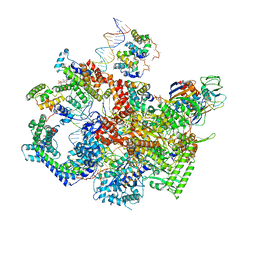 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | 分子名称: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | 著者 | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | 登録日 | 2020-04-21 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.43 Å) | | 主引用文献 | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6K4Y
 
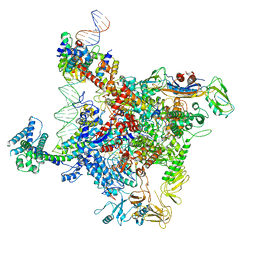 | | CryoEM structure of sigma appropriation complex | | 分子名称: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2019-05-27 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
4ZH3
 
 | |
4ZH4
 
 | |
4ZH2
 
 | |
8IGR
 
 | | Cryo-EM structure of CII-dependent transcription activation complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | 登録日 | 2023-02-21 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8IGS
 
 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | 登録日 | 2023-02-21 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8JO2
 
 | | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA | | 分子名称: | DNA (65-MER), DNA-binding transcriptional regulator BasR, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Lou, Y.-C, Huang, H.-Y, Chen, C, Wu, K.-P. | | 登録日 | 2023-06-06 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA.
Nucleic Acids Res., 51, 2023
|
|
