7YP9
 
 | |
7CHW
 
 | |
6X4Y
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-24 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X43
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | 登録日 | 2020-05-22 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
5UAQ
 
 | | Escherichia coli RNA polymerase RpoB H526Y mutant | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | 登録日 | 2016-12-19 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5IPL
 
 | | SigmaS-transcription initiation complex with 4-nt nascent RNA | | 分子名称: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Zuo, Y, Steitz, T.A. | | 登録日 | 2016-03-09 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structures of E. coli sigma S-transcription initiation complexes provide new insights into polymerase mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6RH3
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-18 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6BYU
 
 | | X-ray crystal structure of Escherichia coli RNA polymerase (RpoB-H526Y) and ppApp complex | | 分子名称: | (5R)-5-(6-amino-9H-purin-9-yl)-2-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}methyl)-4-oxo-4,5-dihydrofuran-3-yl trihydrogen diphosphate, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Murakami, K.S, Molodtsov, V. | | 登録日 | 2017-12-21 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro.
Biochim. Biophys. Acta, 1861, 2018
|
|
6FLQ
 
 | |
7XUI
 
 | |
7KHI
 
 | | Escherichia coli RNA polymerase and rrnBP1 promoter complex with DksA/ppGpp | | 分子名称: | CHAPSO, DNA (28-MER), DNA (36-MER), ... | | 著者 | Shin, Y, Qayyum, M.Z, Murakami, K.S. | | 登録日 | 2020-10-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structural basis of ribosomal RNA transcription regulation.
Nat Commun, 12, 2021
|
|
7DY6
 
 | |
7C97
 
 | |
6N60
 
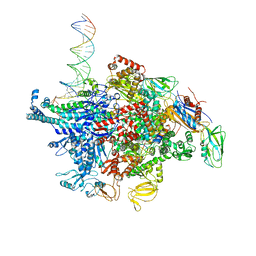 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | 登録日 | 2018-11-23 | | 公開日 | 2019-01-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.68 Å) | | 主引用文献 | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4KN7
 
 | |
6LDI
 
 | |
6GOV
 
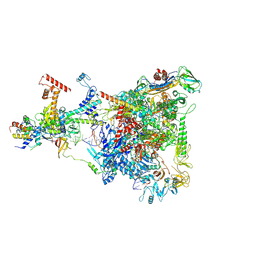 | | Structure of THE RNA POLYMERASE LAMBDA-BASED ANTITERMINATION COMPLEX | | 分子名称: | 30S ribosomal protein S10, Antitermination protein N, DNA (I), ... | | 著者 | Loll, B, Krupp, F, Said, N, Huang, Y, Buerger, J, Mielke, T, Spahn, C.M.T, Wahl, M.C. | | 登録日 | 2018-06-04 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Mol.Cell, 74, 2019
|
|
6GFW
 
 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-02 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
7MKE
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Saecker, R.M, Darst, S.A, Chen, J. | | 登録日 | 2021-04-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6RIN
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-24 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6ALG
 
 | |
6R9G
 
 | |
8EHA
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out) | | 分子名称: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | 登録日 | 2022-09-14 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PSW
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPo) with TraR and rpsT P2 promoter | | 分子名称: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | 登録日 | 2019-07-13 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
