6XSY
 
 | | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Mueller, L.J, Dunn, M.F. | | Deposit date: | 2020-07-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The external aldimine crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-S377A with inhibitor 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine (KOU) at the beta-site.
To be Published
|
|
7NB9
 
 | | E. coli NfsA with nitrofurantoin | | Descriptor: | 1-[(~{E})-(5-nitrofuran-2-yl)methylideneamino]imidazolidine-2,4-dione, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Day, M.D, Jarrom, D, Grainger, A.I, Parr, R.J, Hyde, E.I, White, S.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN.
Biochem.J., 478, 2021
|
|
6DZO
 
 | | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta-site
To be Published
|
|
6DZ4
 
 | | Crystal structure of Salmonella typhimurium Tryptophan Synthase with sodium ion at the metal coordination site and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of Salmonella typhimurium Tryptophan Synthase with sodium ion at the metal coordination site and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta site
To be Published
|
|
5ACC
 
 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
3IRX
 
 | | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the Non-nucleoside RT Inhibitor (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate. | | Descriptor: | (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methy lbenzothioate, Reverse transcriptase, Reverse transcriptase/ribonuclease H | | Authors: | Ho, W.C, Arnold, E. | | Deposit date: | 2009-08-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the
alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor (E)-S-Methyl
5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate.
J.Med.Chem., 52, 2009
|
|
4CJN
 
 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
1SLJ
 
 | | Solution structure of the S1 domain of RNase E from E. coli | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
4UDU
 
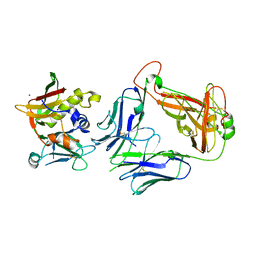 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | ENTEROTOXIN TYPE E, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
6GHN
 
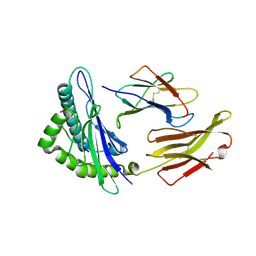 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P9-Phe. | | Descriptor: | ARG-LEU-PRO-ALA-LYS-ALA-PRO-LEU-PHE, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
8V5S
 
 | |
8DAX
 
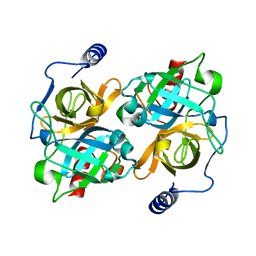 | | New insights into the P186 flip and oligomeric state of Staphylococcus aureus exfoliative toxin E: implications for the exfoliative mechanism | | Descriptor: | Exfoliative toxin E | | Authors: | Gismene, C, Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, Santisteban, A.R.N, de Moraes, F.R, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Staphylococcus aureus Exfoliative Toxin E, Oligomeric State and Flip of P186: Implications for Its Action Mechanism.
Int J Mol Sci, 23, 2022
|
|
1SN8
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1MHE
 
 | | THE HUMAN NON-CLASSICAL MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HLA-E, PEPTIDE (VMAPRTVLL), ... | | Authors: | O'Callaghan, C.A, Tormo, J, Willcox, B.E, Braud, V.B, Jakobsen, B.K, Stuart, D.I, Mcmichael, A.J, Bell, J.I, Jones, E.Y. | | Deposit date: | 1998-08-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E.
Mol.Cell, 1, 1998
|
|
1L5X
 
 | | The 2.0-Angstrom resolution crystal structure of a survival protein E (SurE) homolog from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, Survival protein E | | Authors: | Mura, C, Katz, J.E, Clarke, S.G, Eisenberg, D. | | Deposit date: | 2002-03-08 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of an Archaeal Homolog of Survival
Protein E (SurE-alpha): An Acid Phosphatase with Purine
Nucleotide Specificity
J.Mol.Biol., 326, 2003
|
|
1L5J
 
 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
2VRT
 
 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
1MFP
 
 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
1O6S
 
 | | Internalin (Listeria monocytogenes) / E-Cadherin (human) Recognition Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, E-CADHERIN, ... | | Authors: | Schubert, W.-D, Urbanke, C, Ziehm, T, Beier, V, Machner, M.P, Domann, E, Wehland, J, Chakraborty, T, Heinz, D.W. | | Deposit date: | 2002-10-13 | | Release date: | 2002-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Internalin, a Major Invasion Protein of Listeria Monocytogenes, in Complex with its Human Receptor E-Cadherin
Cell(Cambridge,Mass.), 111, 2002
|
|
2ANQ
 
 | | Crystal Structure of E.coli DHFR in complex with NADPH and the inhibitor compound 10a. | | Descriptor: | (2,5-dimethylbenzene-1,4-diyl)dimethanediyl bis(N-carbamimidoylcarbamimidothioate), Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
3CDG
 
 | | Human CD94/NKG2A in complex with HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Petrie, E.J, Clements, C.S, Lin, J, Sullivan, L.C, Johnson, D, Huyton, T, Heroux, A, Hoare, H.L, Beddoe, T, Reid, H.H, Wilce, M.C.J, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence
J.Exp.Med., 205, 2008
|
|
4ILV
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
4ILT
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | CHLORIDE ION, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2012-12-31 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
2ANO
 
 | | Crystal structure of E.coli dihydrofolate reductase in complex with NADPH and the inhibitor MS-SH08-17 | | Descriptor: | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Summerfield, R.L, Daigle, D.M, Mayer, S, Jackson, S.G, Organ, M, Hughes, D.W, Brown, E.D, Junop, M.S. | | Deposit date: | 2005-08-11 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A 2.13 A Structure of E. coli Dihydrofolate Reductase Bound to a Novel Competitive Inhibitor Reveals a New Binding Surface Involving the M20 Loop Region
J.Med.Chem., 49, 2006
|
|
