6PLG
 
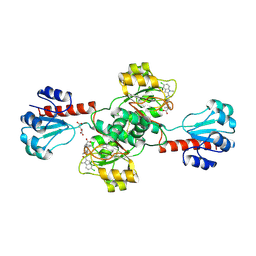 | | Crystal structure of human PHGDH complexed with Compound 15 | | Descriptor: | (2S)-(4-{3-[(4,5-dichloro-1-methyl-1H-indole-2-carbonyl)amino]oxetan-3-yl}phenyl)(pyridin-3-yl)acetic acid, D-3-phosphoglycerate dehydrogenase, D-MALATE | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8HTY
 
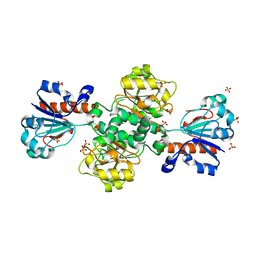 | | Candida boidinii Formate Dehydrogenase Crystal Structure at 1.4 Angstrom Resolution | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Gul, M, Yuksel, B, Bulut, H, DeMirci, H. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of wild-type and Val120Thr mutant Candida boidinii formate dehydrogenase by X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1QP8
 
 | |
5J23
 
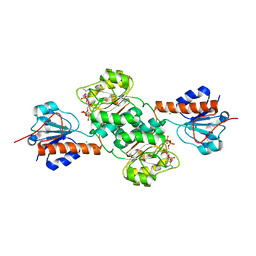 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with 2'-phospho-ADP-ribose | | Descriptor: | 2-hydroxyacid dehydrogenase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Kutner, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
6WKW
 
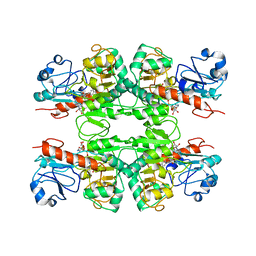 | |
8OQ2
 
 | | Binding of NADP to a formate dehydrogenase from Starkeya novella. | | Descriptor: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Partipilo, M, Whittaker, J.J, Pontillo, N, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of NADP to a formate dehydrogenase from Starkeya novella.
To Be Published
|
|
7ARZ
 
 | | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Goryaynova, D.A, Nikolaeva, A.Y, Pometun, A.A, Savin, S.S, Parshin, P.D, Popov, V.O, Tishkov, V.I, Boyko, K.M. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ternary complex of NAD-dependent formate dehydrogenase from Physcomitrium patens
To Be Published
|
|
7QZ1
 
 | | Formate dehydrogenase from Starkeya novella | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Pontillo, N, Slotboom, D.J, Guskov, A. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural insight into the chemical resistance and cofactor specificity of the formate dehydrogenase from Starkeya novella.
Febs J., 290, 2023
|
|
8GRV
 
 | |
1SC6
 
 | |
3OET
 
 | | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Edwards, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | D-Erythronate-4-Phosphate Dehydrogenase complexed with NAD
To be Published
|
|
7CVP
 
 | | The Crystal Structure of human PHGDH from Biortus. | | Descriptor: | D-3-phosphoglycerate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Miao, Q, Huang, Y. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of human PHGDH from Biortus.
To Be Published
|
|
1DXY
 
 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
3DC2
 
 | |
3DDN
 
 | |
4LCE
 
 | | CtBP1 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
2W2L
 
 | | Crystal structure of the holo forms of Rhodotorula graminis D- mandelate dehydrogenase at 2.5A. | | Descriptor: | D-MANDELATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
2W2K
 
 | | Crystal structure of the apo forms of Rhodotorula graminis D- mandelate dehydrogenase at 1.8A. | | Descriptor: | D-MANDELATE DEHYDROGENASE | | Authors: | Vachieri, S.G, Cole, A.R, Bagneris, C, Baker, D.P, Fewson, C.A, Basak, A.K. | | Deposit date: | 2008-11-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Apo and Holo Forms of Rhodotorula Graminis D(-)-Mandelate Dehydrogenase
To be Published
|
|
7DKM
 
 | | PHGDH covalently linked to oridonin | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CHLORIDE ION, D-3-phosphoglycerate dehydrogenase, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors.
Cell.Mol.Life Sci., 79, 2021
|
|
4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
4CUK
 
 | |
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
4CUJ
 
 | |
3WNV
 
 | | Crystal structure of a glyoxylate reductase from Paecilomyes thermophila | | Descriptor: | SULFATE ION, glyoxylate reductase | | Authors: | Duan, X, Hu, S, Zhou, P, Zhou, Y, Jiang, Z. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a first fungal glyoxylate reductase from Paecilomyes thermophila
Enzyme.Microb.Technol., 60, 2014
|
|
1PSD
 
 | |
