1YCS
 
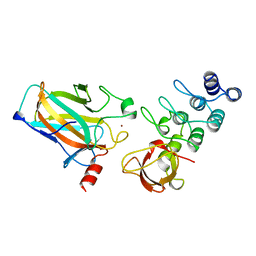 | | P53-53BP2 COMPLEX | | Descriptor: | 53BP2, P53, ZINC ION | | Authors: | Gorina, S, Pavletich, N.P. | | Deposit date: | 1996-09-30 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2.
Science, 274, 1996
|
|
1AWC
 
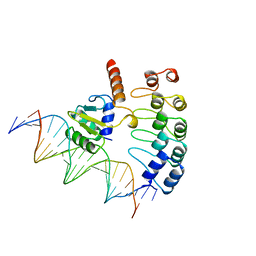 | | MOUSE GABP ALPHA/BETA DOMAIN BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*(BRU)P*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*AP*(CBR)P*AP*CP*(CBR)P*GP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*GP*(BRU)P*GP*(BRU)P*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*AP*T)-3'), PROTEIN (GA BINDING PROTEIN ALPHA), ... | | Authors: | Batchelor, A.H, Wolberger, C. | | Deposit date: | 1997-10-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA.
Science, 279, 1998
|
|
1AP7
 
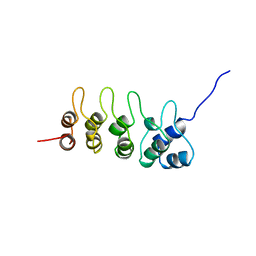 | | P19-INK4D FROM MOUSE, NMR, 20 STRUCTURES | | Descriptor: | P19-INK4D | | Authors: | Archer, S.J, Luh, F.Y, Domaille, P.J, Smith, B.O, Laue, E.D. | | Deposit date: | 1997-07-25 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the cyclin-dependent kinase inhibitor p19Ink4d.
Nature, 389, 1997
|
|
1NFI
 
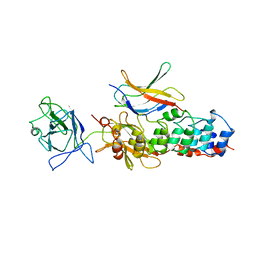 | | I-KAPPA-B-ALPHA/NF-KAPPA-B COMPLEX | | Descriptor: | I-KAPPA-B-ALPHA, NF-KAPPA-B P50, NF-KAPPA-B P65 | | Authors: | Jacobs, M.D, Harrison, S.C. | | Deposit date: | 1998-08-25 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an IkappaBalpha/NF-kappaB complex.
Cell(Cambridge,Mass.), 95, 1998
|
|
1IKN
 
 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
1SW6
 
 | |
1D9S
 
 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
1K1A
 
 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1K1B
 
 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1K3Z
 
 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
1N11
 
 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
1OY3
 
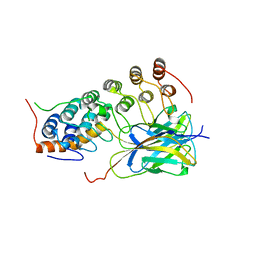 | | CRYSTAL STRUCTURE OF AN IKBBETA/NF-KB P65 HOMODIMER COMPLEX | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Malek, S, Huang, D.B, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
1IXV
 
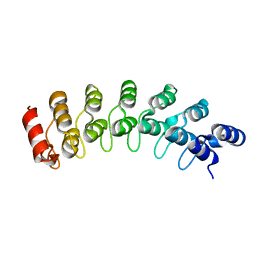 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
1WDY
 
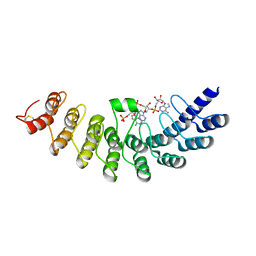 | | Crystal structure of ribonuclease | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Goto, Y, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-05-19 | | Release date: | 2004-10-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of 2',5'-linked oligoadenylates by human ribonuclease L
Embo J., 23, 2004
|
|
1YMP
 
 | |
1WG0
 
 | | Structural comparison of Nas6p protein structures in two different crystal forms | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Nakamura, Y, Umehara, T, Tanaka, A, Horikoshi, M, Yokoyama, S, Padmanabhan, B, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural comparison of Nas6p protein structures in two different crystal forms
To be Published
|
|
1YYH
 
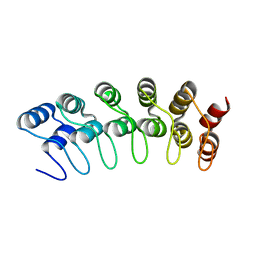 | | Crystal structure of the human Notch 1 ankyrin domain | | Descriptor: | Notch 1, ankyrin domain | | Authors: | Ehebauer, M.T, Chirgadze, D.Y, Hayward, P, Martinez-Arias, A, Blundell, T.L. | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | High-resolution crystal structure of the human Notch 1 ankyrin domain
Biochem.J., 392, 2005
|
|
2FO1
 
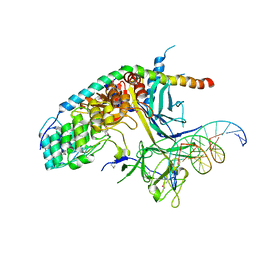 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
2F8Y
 
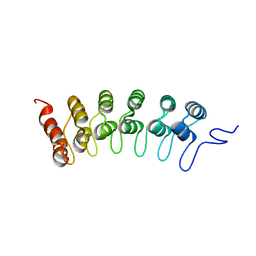 | | Crystal structure of human Notch1 ankyrin repeats to 1.55A resolution. | | Descriptor: | Notch homolog 1, translocation-associated (Drosophila), SULFATE ION | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
2F8X
 
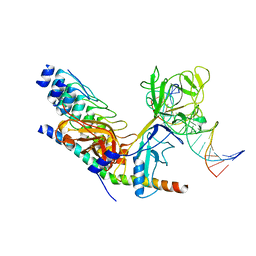 | | Crystal structure of activated Notch, CSL and MAML on HES-1 promoter DNA sequence | | Descriptor: | 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
2HE0
 
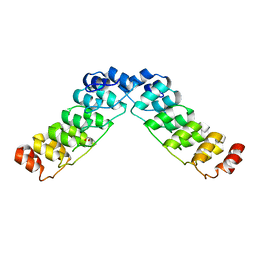 | | Crystal structure of a human Notch1 ankyrin domain mutant | | Descriptor: | 1,2-ETHANEDIOL, Notch1 preproprotein variant | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Martinez Arias, A, Blundell, T.L. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human Notch1 ankyrin domain mutant
TO BE PUBLISHED
|
|
2NYJ
 
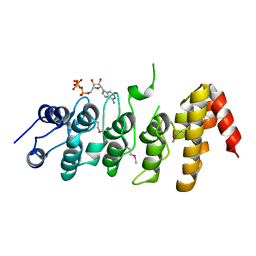 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
2PNN
 
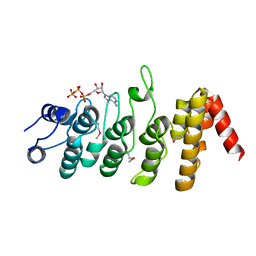 | | Crystal Structure of the Ankyrin Repeat Domain of Trpv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2007-04-24 | | Release date: | 2007-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
2DZN
 
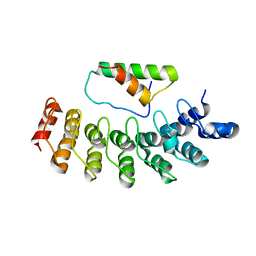 | |
