7OVW
 
 | | Binding domain of botulinum neurotoxin E in complex with GD1a | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin type E | | Authors: | Masuyer, G, Stenmark, P. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Ganglioside Receptor Recognition by Botulinum Neurotoxin Serotype E.
Int J Mol Sci, 22, 2021
|
|
1F24
 
 | |
1O07
 
 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | Descriptor: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | Authors: | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | Deposit date: | 2003-02-20 | | Release date: | 2003-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
1NZ7
 
 | | POTENT, SELECTIVE INHIBITORS OF PROTEIN TYROSINE PHOSPHATASE 1B USING A SECOND PHOSPHOTYROSINE BINDING SITE, complexed with compound 19. | | Descriptor: | 2-[(4-{2-ACETYLAMINO-2-[4-(1-CARBOXY-3-METHYLSULFANYL-PROPYLCARBAMOYL)-BUTYLCARBAMOYL]-ETHYL}-2-ETHYL-PHENYL)-OXALYL-AM INO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Oost, T.K, Abad-Zapatero, C, Hajduk, P.J, Pei, Z, Szczepankiewicz, B.G, Hutchins, C.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Trevillyan, J.M, Jirousek, M.R, Liu, G. | | Deposit date: | 2003-02-16 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent, Selective Inhibitors of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
1SM9
 
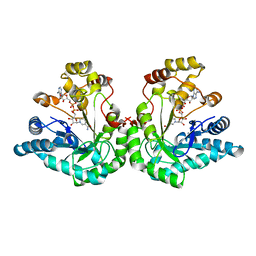 | | Crystal Structure Of An Engineered K274RN276D Double Mutant of Xylose Reductase From Candida Tenuis Optimized To Utilize NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Petschacher, B, Leitgeb, S, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography.
Biochem.J., 385, 2005
|
|
5Y06
 
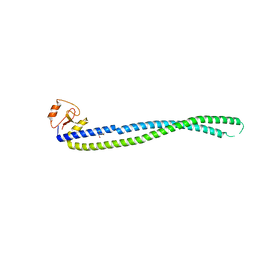 | |
6BMK
 
 | | Crystal structure of MHC-I like protein | | Descriptor: | (2R)-1-(decanoyloxy)-3-(phosphonooxy)propan-2-yl octadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Khandokar, Y.B, Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1SMK
 
 | | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures | | Descriptor: | CITRIC ACID, Malate dehydrogenase, glyoxysomal | | Authors: | Cox, B, Chit, M.M, Weaver, T, Bailey, J, Gietl, C, Bell, E, Banaszak, L. | | Deposit date: | 2004-03-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence.
Febs J., 272, 2005
|
|
1SOA
 
 | | Human DJ-1 with sulfinic acid | | Descriptor: | RNA-binding protein regulatory subunit; oncogene DJ1 | | Authors: | Canet-Aviles, R, Wilson, M.A, Miller, D.W, Ahmad, R, McLendon, C, Bandyopadhyay, S, Baptista, M.J, Ringe, D, Petsko, G.A, Cookson, M.R. | | Deposit date: | 2004-03-13 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1SOW
 
 | |
1F07
 
 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
3P6Q
 
 | |
1SP9
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant Physiol., 134, 2004
|
|
1F25
 
 | |
1EZW
 
 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F3C
 
 | |
1F1A
 
 | | Crystal structure of yeast H48Q cuznsod fals mutant analog | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1NSK
 
 | |
5Y6D
 
 | |
1EZR
 
 | | CRYSTAL STRUCTURE OF NUCLEOSIDE HYDROLASE FROM LEISHMANIA MAJOR | | Descriptor: | CALCIUM ION, NUCLEOSIDE HYDROLASE | | Authors: | Shi, W, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside hydrolase from Leishmania major. Cloning, expression, catalytic properties, transition state inhibitors, and the 2.5-a crystal structure.
J.Biol.Chem., 274, 1999
|
|
1SO7
 
 | | Maltose-induced structure of the human cytolsolic sialidase Neu2 | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
1F10
 
 | |
1ONA
 
 | | CO-CRYSTALS OF CONCANAVALIN A WITH METHYL-3,6-DI-O-(ALPHA-D-MANNOPYRANOSYL)-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bouckaert, J, Maes, D, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1996-07-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-D-mannopyranosyl)-alpha-D-mannopyranoside reveals two binding modes.
J.Biol.Chem., 271, 1996
|
|
1F3P
 
 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | Descriptor: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
1F3H
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
