1U1J
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, METHIONINE, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
3O2M
 
 | |
1U1U
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, SULFATE ION, ZINC ION | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
4PYI
 
 | | human apo COMT | | Descriptor: | Catechol O-methyltransferase, SODIUM ION | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P7F
 
 | | Mouse apo-COMT | | Descriptor: | Catechol O-methyltransferase, HYDROGENPHOSPHATE ION, PHOSPHATE ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OEY
 
 | | Crystal structure of InhA_T266E:NADH complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Molle, V, Gulten, G, Vilcheze, C, Veyron-Churlet, R, Zanella-Cleon, I, Sacchettini, J.C, Jacobs Jr, W.R, Kremer, L. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis.
Mol.Microbiol., 78, 2010
|
|
3OEW
 
 | | Crystal structure of wild-type InhA:NADH complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Molle, V, Gulten, G, Vilcheze, C, Veyron-Churlet, R, Zanella-Cleon, I, Sacchettini, J.C, Jacobs Jr, W.R, Kremer, L. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis.
Mol.Microbiol., 78, 2010
|
|
3OF2
 
 | | Crystal structure of InhA_T266D:NADH complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Molle, V, Gulten, G, Vilcheze, C, Veyron-Churlet, R, Zanella-Cleon, I, Sacchettini, J.C, Jacobs Jr, W.R, Kremer, L. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis.
Mol.Microbiol., 78, 2010
|
|
3OTS
 
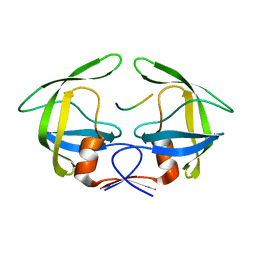 | | MDR769 HIV-1 protease complexed with MA/CA hepta-peptide | | Descriptor: | MA/CA substrate peptide, Multi-drug resistant HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OTY
 
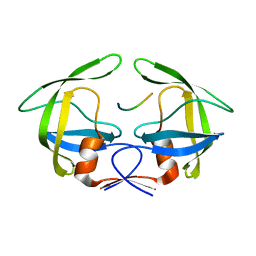 | | MDR769 HIV-1 protease complexed with RT/RH hepta-peptide | | Descriptor: | MDR HIV-1 protease, RT/RH substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUA
 
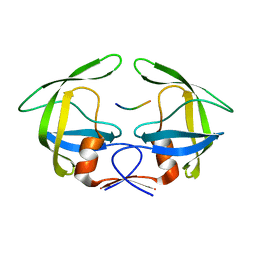 | | MDR769 HIV-1 protease complexed with p1/p6 hepta-peptide | | Descriptor: | HIV-1 protease, p1/p6 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3P2B
 
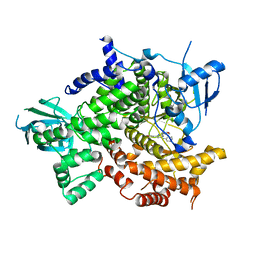 | | Crystal Structure of PI3K gamma with 3-(2-morpholino-6-(pyridin-3-ylamino)pyrimidin-4-yl)phenol | | Descriptor: | 3-[2-morpholin-4-yl-6-(pyridin-3-ylamino)pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knapp, M.S, Elling, R.A, Ornelas, E. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and structure-activity relationship of 2-morpholino 6-(3-hydroxyphenyl) pyrimidines, a class of potent and selective PI3 kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OU1
 
 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OWK
 
 | | Human CK2 catalytic domain in complex with a benzopyridoindole derivative inhibitor | | Descriptor: | 7-chloro-10-methyl-11H-benzo[g]pyrido[4,3-b]indol-3-ol, CSNK2A1 protein, SULFATE ION | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
3OU4
 
 | | MDR769 HIV-1 protease complexed with TF/PR hepta-peptide | | Descriptor: | HIV-1 protease, TF/PR substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OU3
 
 | | MDR769 HIV-1 protease complexed with PR/RT hepta-peptide | | Descriptor: | HIV-1 protease, PR/RT substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUC
 
 | | MDR769 HIV-1 protease complexed with p2/NC hepta-peptide | | Descriptor: | MDR HIV-1 protease, p2/NC substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OWJ
 
 | | Human CK2 catalytic domain in complex with a pyridocarbazole derivative inhibitor | | Descriptor: | 9-hydroxy-5,11-dimethyl-4,6-dihydro-1H-pyrido[4,3-b]carbazol-1-one, CSNK2A1 protein | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
3OUB
 
 | | MDR769 HIV-1 protease complexed with NC/p1 hepta-peptide | | Descriptor: | MDR HIV-1 protease, NC/p1 substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OUD
 
 | | MDR769 HIV-1 protease complexed with CA/p2 hepta-peptide | | Descriptor: | CA/p2 substrate peptide, MDR HIV-1 protease | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
3OWL
 
 | | Human CK2 catalytic domain in complex with a benzopyridoindole derivative inhibitor | | Descriptor: | 11-chloro-8-methyl-7H-benzo[e]pyrido[4,3-b]indol-3-ol, CSNK2A1 protein, SULFATE ION | | Authors: | Reiser, J.-B, Prudent, R, Cochet, C. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antitumor activity of pyridocarbazole and benzopyridoindole derivatives that inhibit protein kinase CK2.
Cancer Res., 70, 2010
|
|
4PHU
 
 | | Crystal structure of Human GPR40 bound to allosteric agonist TAK-875 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIMETHYL SULFOXIDE, Free fatty acid receptor 1,Lysozyme, ... | | Authors: | Srivastava, A, Yano, J.K, Hirozane, Y, Kefala, G, Snell, G, Lane, W, Gruswitz, F, Ivetac, A, Aertgeerts, K, Nguyen, J, Jennings, A, Okada, K. | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875.
Nature, 513, 2014
|
|
4P5E
 
 | | CRYSTAL STRUCTURE OF HUMAN DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, CALCIUM ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
2F6G
 
 | | BenM effector binding domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Clark, T, Haddad, S, Ezezika, O, Neidle, E, Momany, C. | | Deposit date: | 2005-11-29 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Distinct Effector-binding Sites Enable Synergistic Transcriptional Activation by BenM, a LysR-type Regulator.
J.Mol.Biol., 367, 2007
|
|
4P7K
 
 | | Rat COMT in complex with sinefungin | | Descriptor: | ACETATE ION, Catechol O-methyltransferase, L(+)-TARTARIC ACID, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
