3LLM
 
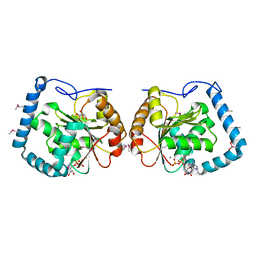 | | Crystal Structure Analysis of a RNA Helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, CACODYLATE ION, ... | | Authors: | Schutz, P, Karlberg, T, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein.
J.Mol.Biol., 400, 2010
|
|
6XU6
 
 | | Drosophila melanogaster Testis 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU8
 
 | | Drosophila melanogaster Ovary 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU7
 
 | | Drosophila melanogaster Testis polysome ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
3MEK
 
 | | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Lam, R, Dombrovski, L, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine
To be Published
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8H3Q
 
 | | Cryo-EM Structure of the CAND1-Cul3-Rbx1 complex | | Descriptor: | Cullin-3, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ETS
 
 | | Class1 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETU
 
 | | Class2 of the INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETW
 
 | | Class3 of INO80-Hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
6K8E
 
 | | Global regulatory element SarX | | Descriptor: | HTH-type transcriptional regulator SarX | | Authors: | Wang, Q. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of SarX from Staphylococcus aureus
To Be Published
|
|
4N9I
 
 | | Crystal Structure of Transcription regulation protein CRP complexed with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Catabolite gene activator | | Authors: | Lee, B.-J, Seok, S.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5AQC
 
 | | KstR, transcriptional repressor of cholesterol degradation in Mycobacterium tuberculosis, bound to the cholesterol coenzyme A derivative, (25R)-3-oxocholest-4-en-26-oyl-CoA. | | Descriptor: | (25S)-3-oxocholest-4-en-26-oyl-CoA, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Podust, L.M, Ouellet, H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kstr, Transcriptional Repressor of Cholesterol Degradation in Mycobacterium Tuberculosis, Bound to the Cholesterol Coenzyme a Derivative, (25S)-3- Oxocholest-4-En-26-Oyl-Coa.
To be Published
|
|
1GL6
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolysable GTP analogue GDPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATPASE, CHLORIDE ION, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GKI
 
 | | Plasmid coupling protein TrwB in complex with ADP and Mg2+. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CONJUGAL TRANSFER PROTEIN TRWB, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
5A5U
 
 | | Structure of mammalian eIF3 in the context of the 43S preinitiation complex | | Descriptor: | EUKARYOTIC INITIATION FACTOR 3, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT B, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT I | | Authors: | des-Georges, A, Dhote, V, Kuhn, L, Hellen, C.U.T, Pestova, T.V, Frank, J, Hashem, Y. | | Deposit date: | 2015-06-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of Mammalian Eif3 in the Context of the 43S Preinitiation Complex.
Nature, 525, 2015
|
|
4ZS7
 
 | | Structural mimicry of receptor interaction by antagonistic IL-6 antibodies | | Descriptor: | Interleukin-6, Llama Fab fragment 68F2 heavy chain, Llama Fab fragment 68F2 light chain | | Authors: | Blanchetot, C, De Jonge, N, Desmyter, A, Ongenae, N, Hofman, E, Klarenbeek, A, Sadi, A, Hultberg, A, Kretz-Rommel, A, Spinelli, S, Loris, R, Cambillau, C, de Haard, H. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural Mimicry of Receptor Interaction by Antagonistic Interleukin-6 (IL-6) Antibodies.
J.Biol.Chem., 291, 2016
|
|
5WG6
 
 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
1E9R
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Trigonal form in complex with sulphate. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB, SULFATE ION | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1E9S
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Unbound monoclinic form. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GL7
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolisable ATP-analogue ADPNP. | | Descriptor: | CHLORIDE ION, CONJUGAL TRANSFER PROTEIN TRWB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
4N9H
 
 | | Crystal structure of Transcription regulation Protein CRP | | Descriptor: | Catabolite gene activator | | Authors: | Lee, B.J, Seok, S.H, Im, H, Yoon, H.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
