9EC2
 
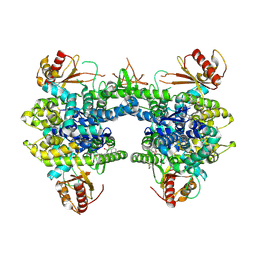 | | Crystal structure of SAMHD1 dimer bound to an inhibitor obtained from high-throughput chemical tethering to the guanine antiviral acyclovir | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, N-[5-({2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl}amino)-5-oxopentyl]-4,7-dibromo-3-hydroxynaphthalene-2-carboxamide | | Authors: | Egleston, M, Dong, L, Howlader, A.H, Bhat, S, Orris, B, Lopez-Rovira, L.M, Bianchet, M.A, Greenberg, M.M, Stivers, J.T. | | Deposit date: | 2024-11-13 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Inhibitors of SAMHD1 Obtained from Chemical Tethering to the Guanine Antiviral Acyclovir.
Biochemistry, 64, 2025
|
|
6Q96
 
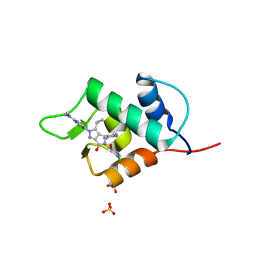 | |
9MGP
 
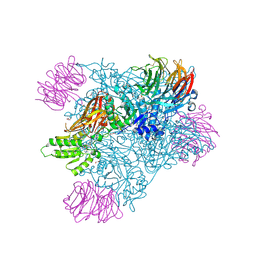 | |
5GYB
 
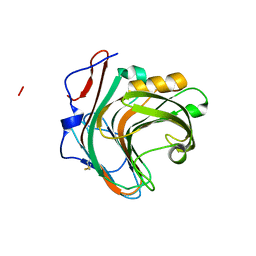 | |
7SZP
 
 | |
9GC5
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | 1-[2-[5-(1,3-benzoxazol-4-yl)-2,4-bis(fluoranyl)phenyl]-3-pyrimidin-4-yl-4,6-dihydropyrrolo[3,4-d]imidazol-5-yl]propan-1-one, Epidermal growth factor receptor, SULFATE ION | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9MGR
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and compound 51 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 6-[(1-acetylazetidin-3-yl)amino]-N-[(2R)-2-hydroxy-2-{(3S)-7-[(4-methyl-1,3-oxazol-5-yl)methoxy]-1,2,3,4-tetrahydroisoquinolin-3-yl}ethyl]-2-(4-methylpiperidin-1-yl)pyrimidine-4-carboxamide, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | MTA-Cooperative PRMT5 Inhibitors: Mechanism Switching Through Structure-Based Design.
J.Med.Chem., 68, 2025
|
|
8AQ3
 
 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8VTU
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-66, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 1-cyclopropyl-7-[(4-{[3-({(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-14-[({3-[5-(methylcarbamoyl)pyridin-3-yl]prop-2-yn-1-yl}oxy)imino]-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl}oxy)-3-oxopropyl]amino}butyl)amino]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6I6Z
 
 | |
6BSU
 
 | | Crystal structure of xyloglucan xylosyltransferase I | | Descriptor: | MANGANESE (II) ION, Xyloglucan 6-xylosyltransferase 1 | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6PTX
 
 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
5CS6
 
 | | Crystal Structure of CK2alpha with Compound 3 bound | | Descriptor: | 1-(3-chloro-4-propoxyphenyl)methanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8P5B
 
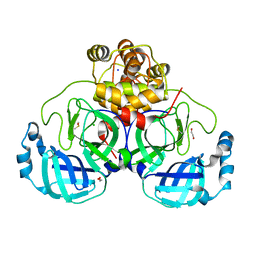 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6BYA
 
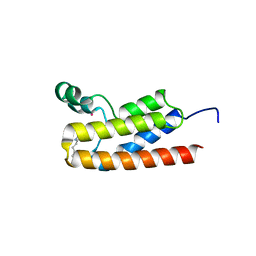 | | Crystal structure of LdBPK_091320 with inhibitor bound | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, UNKNOWN ATOM OR ION, Uncharacterized protein | | Authors: | Dong, A, Lin, Y.H, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of LdBPK_091320.1 with with inhibitor bound
to be published
|
|
8WMD
 
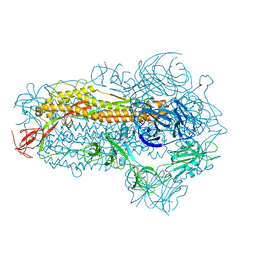 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
7U04
 
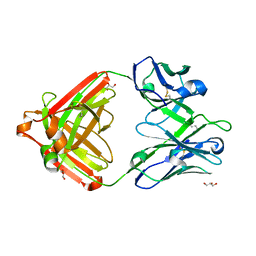 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
6ZPS
 
 | |
9BFV
 
 | | Tri-complex of Compound-23, KRAS G12C, and CypA | | Descriptor: | (3R)-1-[(2E)-4-(dimethylamino)-4-methylpent-2-enoyl]-N-[(2S)-1-{[(1P,8R,10R,14S,21M)-22-ethyl-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methylpyrrolidine-3-carboxamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Elironrasib (RMC-6291), a Potent and Orally Bioavailable, RAS(ON) G12C-Selective, Covalent Tricomplex Inhibitor for the Treatment of Patients with RAS G12C-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
5LL4
 
 | |
7TI2
 
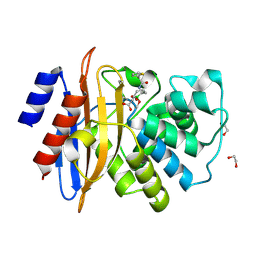 | | Structure of KPC-2 bound to RPX-7063 at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolyzing beta-lactamase KPC, {(3R,7S)-2-hydroxy-3-[2-(thiophen-2-yl)acetamido]-2,3,4,7-tetrahydro-1,2-oxaborepin-7-yl}acetic acid | | Authors: | Clifton, M.C, Fairman, J.W, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
5GY9
 
 | |
2TPS
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | MAGNESIUM ION, PROTEIN (THIAMIN PHOSPHATE SYNTHASE), PYROPHOSPHATE 2-, ... | | Authors: | Chiu, H.-J, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-03-09 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
Biochemistry, 38, 1999
|
|
