1VPR
 
 | |
1VWB
 
 | | STREPTAVIDIN-CYCLO-AC-[CHPQFC]-NH2, PH 11.8 | | Descriptor: | PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
1W7L
 
 | | Crystal structure of human kynurenine aminotransferase I | | Descriptor: | KYNURENINE--OXOGLUTARATE TRANSAMINASE I, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Han, Q, Li, J, Li, J, Rizzi, M. | | Deposit date: | 2004-09-06 | | Release date: | 2004-09-08 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Kynurenine Aminotransferase I
J.Biol.Chem., 279, 2004
|
|
7TV4
 
 | | Crystal structure of NEMO CoZi in complex with HOIP NZF1 and linear diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RNF31, NF-kappa-B essential modulator, Polyubiquitin-C, ... | | Authors: | Rahighi, S, Iyer, M, Oveisi, H. | | Deposit date: | 2022-02-03 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for the simultaneous recognition of NEMO and acceptor ubiquitin by the HOIP NZF1 domain.
Sci Rep, 12, 2022
|
|
8UWX
 
 | |
8UFU
 
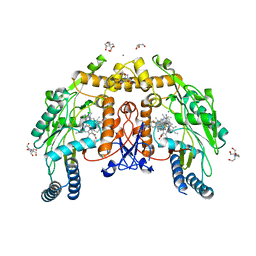 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)-4-methylquinolin-2-amine | | Descriptor: | (7M)-7-[(9S)-9-amino-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl]-4-methylquinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
7TJ7
 
 | |
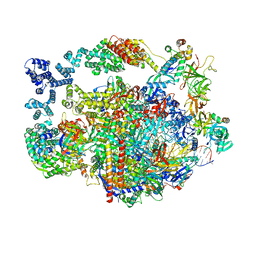
8UHA
 
 | |
8UIS
 
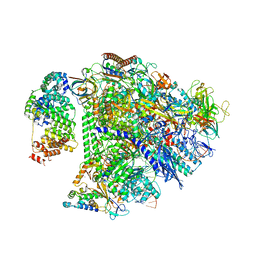 | | Structure of transcription complex Pol II-DSIF-NELF-TFIIS | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB3, DNA-directed RNA polymerase II subunit RPB7, ... | | Authors: | Su, B.G, Vos, S.M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Distinct negative elongation factor conformations regulate RNA polymerase II promoter-proximal pausing.
Mol.Cell, 84, 2024
|
|
7TIJ
 
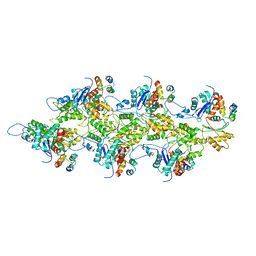 | |
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7U07
 
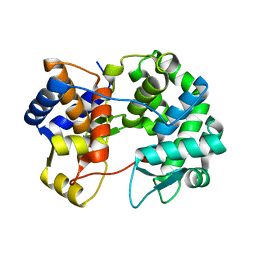 | |
8UZ6
 
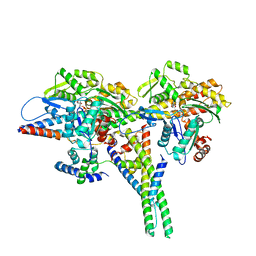 | |
8V0K
 
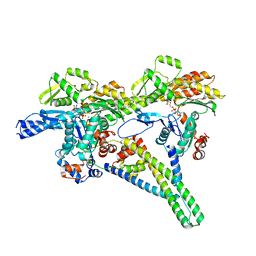 | |
7U91
 
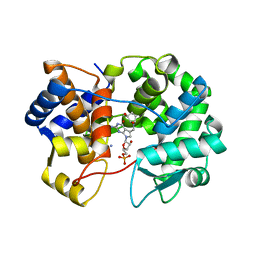 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U1O
 
 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U5A
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant K199C, complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, MALONATE ION, Queuine salvage enzyme DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
8V0I
 
 | |
8VJC
 
 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
8UWY
 
 | |
7TKW
 
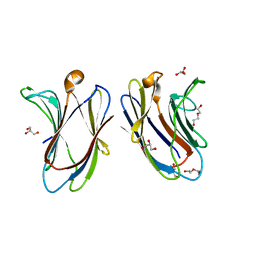 | |
8USX
 
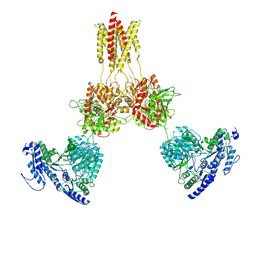 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
8UZ5
 
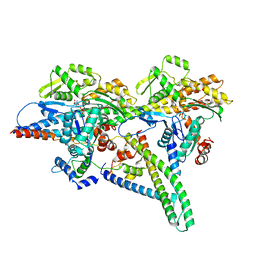 | |
8VJB
 
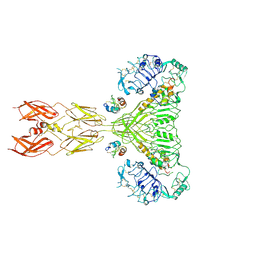 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
7TKX
 
 | | Crystal structure of R14A human Galectin-7 mutant in presence of 4-O-beta-D-Galactopyranosyl-D-glucose | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,3,5, 6-tetrakis(oxidanyl)hexanal, 1,2-ETHANEDIOL, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of R14A human Galectin-7 mutant in presence of 4-O-beta-D-Galactopyranosyl-D-glucose
To Be Published
|
|
