3STV
 
 | |
3DWO
 
 | | Crystal structure of a Pseudomonas aeruginosa FadL homologue | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Probable outer membrane protein, SULFATE ION | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2008-07-22 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall.
Nature, 458, 2009
|
|
3DWN
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant A77E/S100R | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2008-07-22 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall.
Nature, 458, 2009
|
|
6CYQ
 
 | | Crystal structure of CTX-M-14 S70G/N106S beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
4RLB
 
 | |
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
7LH5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with plazomicin, mRNA and tRNAs | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Golkar, T, Berghuis, A.M, Schmeing, T.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
4S2W
 
 | | Structure of E. coli RppH bound to sulfate ions | | Descriptor: | RNA pyrophosphohydrolase, SULFATE ION | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
6CYK
 
 | | CTX-M-14 N106S mutant | | Descriptor: | ACETATE ION, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
6CYN
 
 | | CTX-M-14 N106S/D240G mutant | | Descriptor: | Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
4RL9
 
 | |
4RLC
 
 | |
6CYU
 
 | | Crystal structure of CTX-M-14 S70G/N106S/D240G beta-lactamase in complex with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Patel, M.P, Hu, L, Sankaran, B, Brown, C, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2018-04-06 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synergistic effects of functionally distinct substitutions in beta-lactamase variants shed light on the evolution of bacterial drug resistance.
J. Biol. Chem., 293, 2018
|
|
4S2Y
 
 | | Structure of E. coli RppH bound to RNA and three magnesium ions | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4S2V
 
 | | E. coli RppH structure, KI soak | | Descriptor: | ACETATE ION, CALCIUM ION, IODIDE ION, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4TWI
 
 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
3EIG
 
 | |
4RUB
 
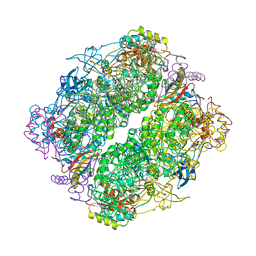 | | A CRYSTAL FORM OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE FROM NICOTIANA TABACUM IN THE ACTIVATED STATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Schreuder, H, Cascio, D, Curmi, P.M.G, Eisenberg, D. | | Deposit date: | 1990-05-25 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal form of ribulose-1,5-bisphosphate carboxylase/oxygenase from Nicotiana tabacum in the activated state.
J.Mol.Biol., 197, 1987
|
|
4S2X
 
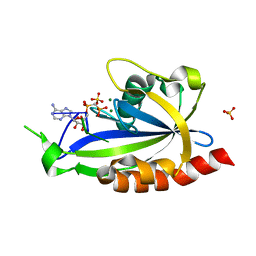 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
4TWJ
 
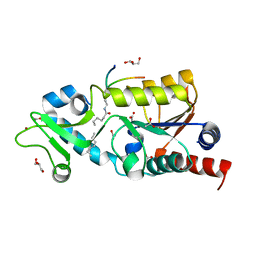 | | The structure of Sir2Af2 bound to a myristoylated histone peptide | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4 peptide, ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4U3C
 
 | | Docking Site of Maltohexaose in the Mtb GlgE | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
6U6D
 
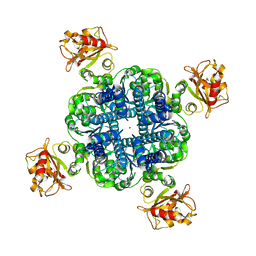 | | MthK closed state with EDTA | | Descriptor: | Calcium-gated potassium channel MthK, POTASSIUM ION | | Authors: | Fan, C, Rheinberger, J, Nimigean, C.M. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
4U2Y
 
 | | Sco GlgEI-V279S in Complex with Reaction Intermediate Azasugar | | Descriptor: | (2R,3R,4R,5R)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-3-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1 | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U31
 
 | | Sco GlgEI-V279S in Complex with maltose-C-phosphonate | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
4U33
 
 | | Structure of Mtb GlgE bound to maltose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.293 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis GlgE and complexes with non-covalent inhibitors.
Sci Rep, 5, 2015
|
|
