1AWV
 
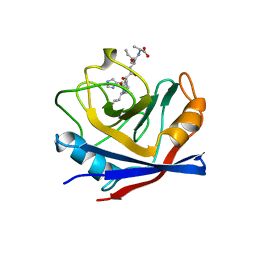 | | CYPA COMPLEXED WITH HVGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1XCZ
 
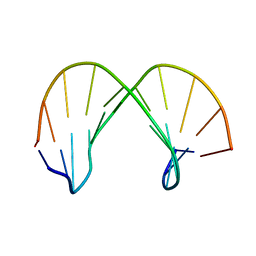 | | Structure of DNA containing the beta-anomer of a carbocyclic abasic site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(DXD)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, El-khateeb, M, Rege, P, Tian, K, Johnson, F. | | Deposit date: | 2004-09-03 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of the C1 configuration of abasic sites on DNA duplex structure
Biochemistry, 43, 2004
|
|
1PD9
 
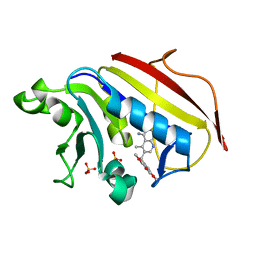 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1BE9
 
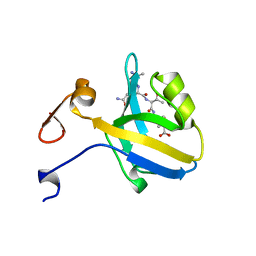 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1B9R
 
 | | TERPREDOXIN FROM PSEUDOMONAS SP. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (TERPREDOXIN) | | Authors: | Mo, H, Pochapsky, S.S, Pochapsky, T.C. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas.
Biochemistry, 38, 1999
|
|
5TTB
 
 | |
1BH0
 
 | | STRUCTURE OF A GLUCAGON ANALOG | | Descriptor: | GLUCAGON | | Authors: | Sturm, N.S, Lin, Y, Burley, S.K, Krstenansky, J.L, Ahn, J.-M, Azizeh, B.Y, Trivedi, D, Hruby, V.J. | | Deposit date: | 1998-06-11 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies on positions 17, 18, and 21 replacement analogues of glucagon: the importance of charged residues and salt bridges in glucagon biological activity.
J.Med.Chem., 41, 1998
|
|
5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
5UCE
 
 | | Solution NMR structure of the major species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
3Q8C
 
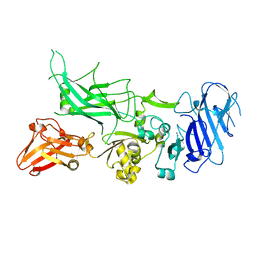 | | Crystal structure of Protective Antigen W346F (pH 5.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
5UB0
 
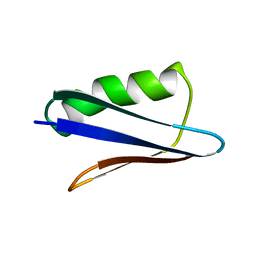 | | Solution NMR Structure of NERD-C, a natively folded tetramutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
1JQ1
 
 | |
5UCF
 
 | | Solution NMR-derived model of the minor species of DANCER-2, a dynamic and natively folded pentamutant of the B1 domain of streptococcal protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Davey, J.A, Goto, N.K, Chica, R.A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational design of proteins that exchange on functional timescales.
Nat. Chem. Biol., 13, 2017
|
|
6SE6
 
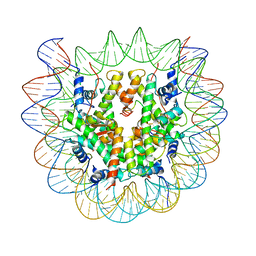 | | Class2 : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
5G5J
 
 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | Descriptor: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | Deposit date: | 2013-01-23 | | Release date: | 2013-12-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
5JUL
 
 | | Near atomic structure of the Dark apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Cheng, T.C, Akey, I.V, Yuan, S, Yu, Z, Ludtke, S.J, Akey, C.W. | | Deposit date: | 2016-05-10 | | Release date: | 2017-02-22 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation.
Structure, 25, 2017
|
|
5GZR
 
 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
1BGB
 
 | | ECORV ENDONUCLEASE COMPLEX WITH 5'-CGGGATATCCC DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), ECORV ENDONUCLEASE | | Authors: | Perona, J, Horton, N.C. | | Deposit date: | 1998-05-28 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of flanking DNA sequences by EcoRV endonuclease involves alternative patterns of water-mediated contacts.
J.Biol.Chem., 273, 1998
|
|
1X7K
 
 | |
6SEE
 
 | | Class2A : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
1B9P
 
 | |
5G6U
 
 | | Crystal structure of langerin carbohydrate recognition domain with GlcNS6S | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Porkolab, V, Chabrol, E, Varga, N, Ordanini, S, Sutkeviciute, I, Thepaut, M, Bernardi, A, Fieschi, F. | | Deposit date: | 2016-07-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Rational-Differential Design of Highly Specific Glycomimetic Ligands: Targeting DC-SIGN and Excluding Langerin Recognition.
ACS Chem. Biol., 13, 2018
|
|
1O00
 
 | |
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
