2D2K
 
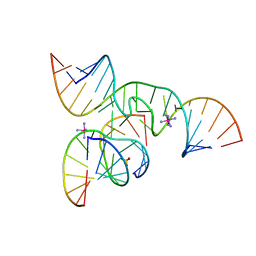 | | Crystal Structure of a minimal, native (U39) all-RNA hairpin ribozyme | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
2D2L
 
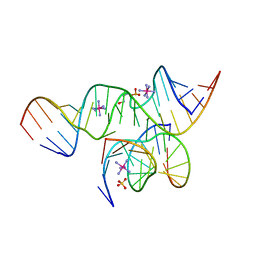 | | Crystal Structure of a minimal, all-RNA hairpin ribozyme with a propyl linker (C3) at position U39 | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
6WTL
 
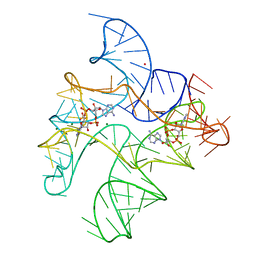 | | Structure of Human pir-miRNA-19b-2 Apical Loop and One-base-pair Fused to the YdaO Riboswitch Scaffold | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Shoffner, G.M, Peng, Z, Guo, F. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
7LYJ
 
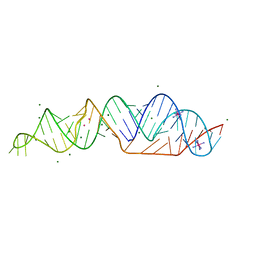 | | SARS-CoV-2 frameshifting pseudoknot RNA | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2021-03-07 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot.
Rna, 28, 2022
|
|
7MKY
 
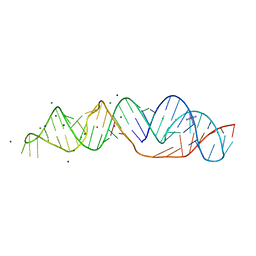 | | SARS-CoV-2 frameshifting pseudoknot RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT HEXAMMINE(III), MAGNESIUM ION, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot.
Rna, 28, 2022
|
|
2GIC
 
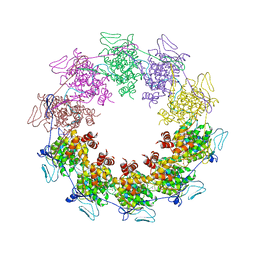 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
4Q4Z
 
 | | Thermus thermophilus RNA polymerase de novo transcription initiation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ADENOSINE-5'-TRIPHOSPHATE, DNA (25-MER), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-15 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
4K4U
 
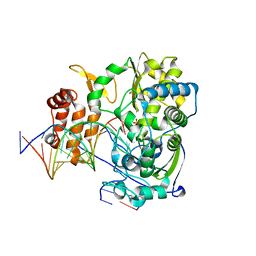 | | Poliovirus polymerase elongation complex (r5_form) | | Descriptor: | RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*A)-3'), RNA (5'-R(P*GP*GP*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
1D16
 
 | |
6EMS
 
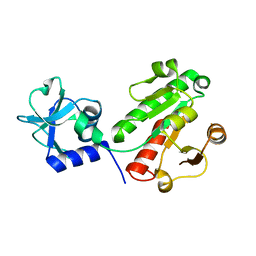 | |
4K4W
 
 | | Poliovirus polymerase elongation complex (r5+2_form) | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*GP*AP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*AP*GP*AP*UP*GP*AP*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*UP*CP*GP*UP*CP*GP*AP*AP*A)-3'), RNA-directed RNA polymerase 3D-POL | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
8YGJ
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
4Q5S
 
 | | Thermus thermophilus RNA polymerase initially transcribing complex containing 6-mer RNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*GP*CP*AP*GP*CP*CP*A)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3'), ... | | Authors: | Murakami, K.S. | | Deposit date: | 2014-04-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme.
J.Biol.Chem., 289, 2014
|
|
6UP0
 
 | |
8YNP
 
 | | RNA duplex containing Methylformamide | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*(5BU)P*CP*GP*AP*GP*(HHX)P*CP*C)-3') | | Authors: | Kondo, J, Abe, H. | | Deposit date: | 2024-03-11 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of 2'-formamidonucleoside phosphoramidites for suppressing the seed-based off-target effects of siRNAs.
Nucleic Acids Res., 52, 2024
|
|
8VFS
 
 | |
8YNO
 
 | | RNA duplex containing Formamide | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*(5BU)P*CP*GP*AP*GP*(HHU)P*CP*C)-3'), SPERMINE | | Authors: | Kondo, J, Abe, H. | | Deposit date: | 2024-03-11 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of 2'-formamidonucleoside phosphoramidites for suppressing the seed-based off-target effects of siRNAs.
Nucleic Acids Res., 52, 2024
|
|
1FO1
 
 | |
4QBT
 
 | |
3ZXI
 
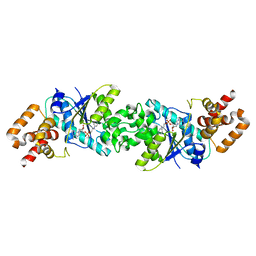 | | Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with a tyrosyl-adenylate analog | | Descriptor: | PHOSPHORIC ACID 2-AMINO-3-(4-HYDROXY-PHENYL)-PROPYL ESTER ADENOSIN-5'YL ESTER, TYROSYL-TRNA SYNTHETASE, MITOCHONDRIAL | | Authors: | Bonnefond, L, Frugier, M, Rudinger-Thirion, J, Balg, C, Chenevert, R, Lorber, B, Giege, R, Sauter, C. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
1L3Z
 
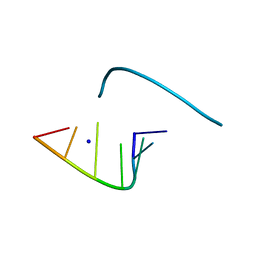 | | Crystal Structure Analysis of an RNA Heptamer | | Descriptor: | 5'-R(*GP*UP*AP*UP*AP*CP*A)-3', SODIUM ION | | Authors: | Shi, K, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-03-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of an alternating RNA heptamer r(GUAUACA)
forming a six base-paired duplex with 3'-end adenine overhangs
Nucleic Acids Res., 31, 2003
|
|
4PMI
 
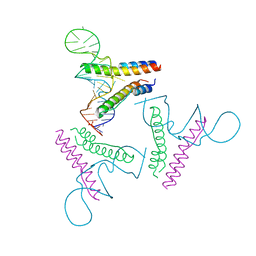 | | Crystal structure of Rev and Rev-response-element RNA complex | | Descriptor: | PHOSPHATE ION, Protein Rev, Rev-Response-Element RNA | | Authors: | Jayaraman, B, Crosby, D.C, Homer, C, Ribeiro, I, Mavor, D, Frankel, A.D. | | Deposit date: | 2014-05-21 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex.
Elife, 4, 2014
|
|
4Q93
 
 | |
4RMO
 
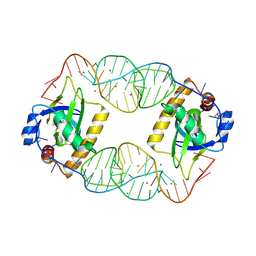 | | Crystal Structure of the CptIN Type III Toxin-Antitoxin System from Eubacterium rectale | | Descriptor: | CALCIUM ION, CptN Toxin, RNA (45-MER) | | Authors: | Rao, F, Voss, J.E, Short, F.L, Luisi, B.F. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system.
Nucleic Acids Res., 43, 2015
|
|
1G1X
 
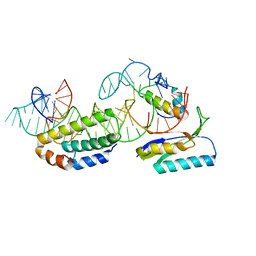 | | STRUCTURE OF RIBOSOMAL PROTEINS S15, S6, S18, AND 16S RIBOSOMAL RNA | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S15, 30S RIBOSOMAL PROTEIN S18, ... | | Authors: | Agalarov, S.C, Prasad, G.S, Funke, P.M, Stout, C.D, Williamson, J.R. | | Deposit date: | 2000-10-13 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain.
Science, 288, 2000
|
|
