6NHR
 
 | |
6NEF
 
 | | Outer Membrane Cytochrome S Filament from Geobacter Sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C, MAGNESIUM ION | | Authors: | Filman, D.J, Marino, S.F, Ward, J.E, Yang, L, Mester, Z, Bullitt, E, Lovley, D.R, Strauss, M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire.
Commun Biol, 2, 2019
|
|
4NDU
 
 | |
6NEN
 
 | | Catalytic domain of Proteus mirabilis ScsC | | Descriptor: | Copper resistance protein | | Authors: | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | Deposit date: | 2018-12-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NHP
 
 | |
6NG3
 
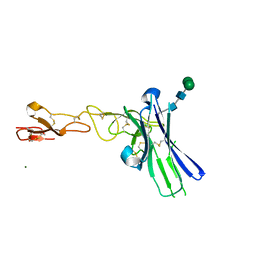 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
4NOF
 
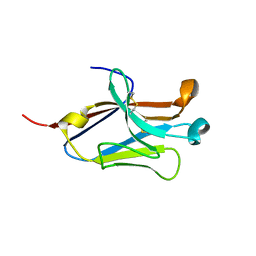 | | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Polymeric immunoglobulin receptor | | Authors: | Sampathkumar, P, Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4NP5
 
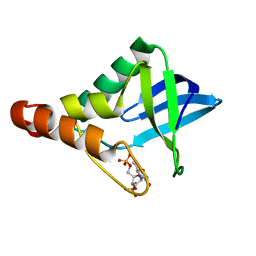 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A/I92N at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
5WQU
 
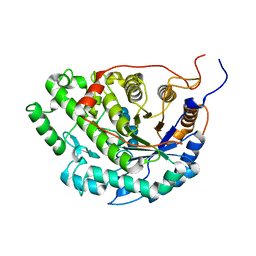 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
5WRR
 
 | | Crystal structure of Fam20A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pseudokinase FAM20A | | Authors: | Zhu, Q. | | Deposit date: | 2016-12-03 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structure of Fam20A reveals a pseudokinase featuring a unique disulfide pattern and inverted ATP-binding
Elife, 6, 2017
|
|
3VGW
 
 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
4NOB
 
 | | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4NCZ
 
 | | Spermidine N-acetyltransferase from Vibrio cholerae in complex with 2-[n-cyclohexylamino]ethane sulfonate. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, SODIUM ION, ... | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-25 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4NMB
 
 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA in complex with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Singh, H, Almo, S.C, Tanner, J.J. | | Deposit date: | 2013-11-14 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NLD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
3M30
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
3E8H
 
 | |
3UL6
 
 | | Saccharum officinarum canecystatin-1 in space group P6422 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Canecystatin-1 | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
3E8V
 
 | | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis | | Descriptor: | Possible transglutaminase-family protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a possible transglutaminase-family protein proteolytic fragment from Bacteroides fragilis
To be Published
|
|
3UOV
 
 | | Crystal Structure of OTEMO (FAD bound form 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.045 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3E93
 
 | |
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | Authors: | Shi, R, Matte, A, Cygler, M, Lau, P. | | Deposit date: | 2011-11-17 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3E9C
 
 | |
4V1I
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 at medium resolution | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3UPL
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | GLYCEROL, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
